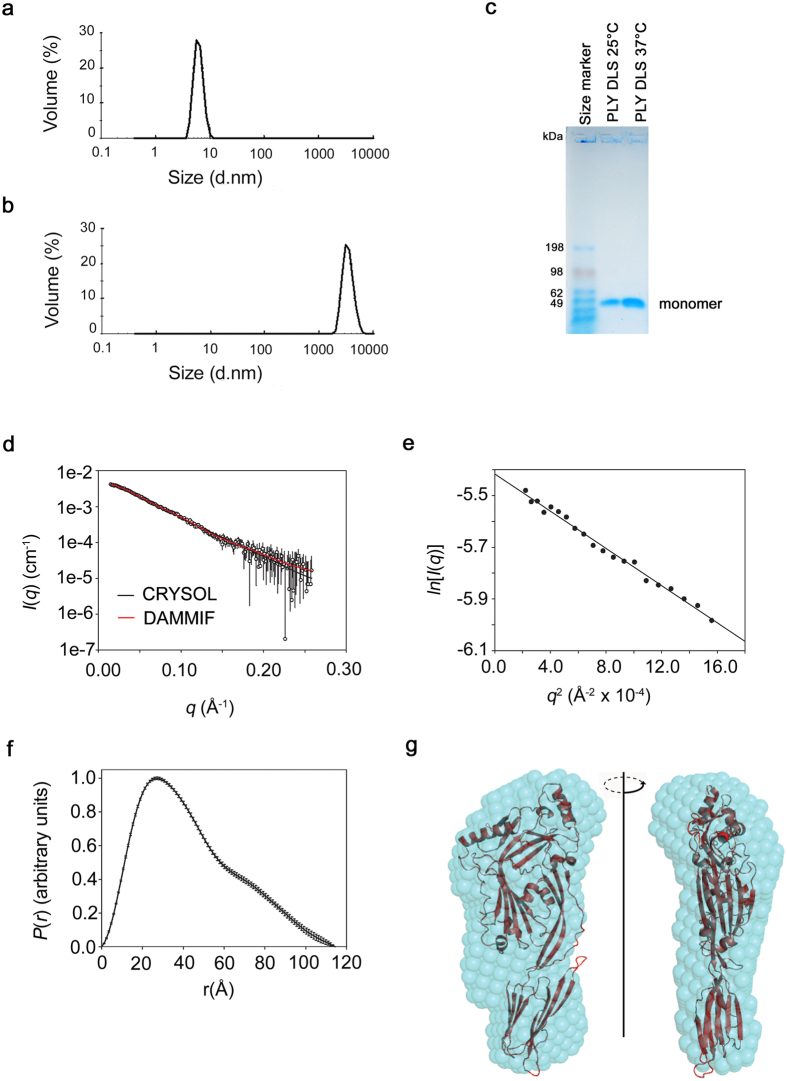
Figure 3. Analysis of PLY oligomers in solution.
(a) Dynamic light scattering profile after incubation at 25 °C. (b) Dynamic light scattering profile after incubation at 37 °C for 10 minutes. (c) SDS agarose gel analysis of the same PLY solutions that were examined by DLS. The expected PLY molecular weight is 53.7 kDa based on amino acid sequence. (d) SEC-SAXS analysis. The mean intensities as a function of the magnitude of the scattering vector (I(q) vs. q) are shown as circles and the error bars indicate ±1 standard deviation. The theoretical scattering profile of the PLY crystal structure was fitted to the experimental data using CRYSOL37 and is shown as a solid black line (ChiCRYSOL = 0.611). The fit of the theoretical scattering profile of a representative dummy atom model generated by DAMMIF36 is shown as a solid red line (ChiDAMMIF = 0.593). (e) Guinier plot. The first 20 data points of the scattering data satisfied q.Rg < 1.3. (f) Pair distance vector distribution function generated from the SAXS data using GNOM63. (g) Comparison of the SAXS-derived ab initio shape envelope with the PLY crystal structure. The averaged filtered shape envelope from DAMAVER53, derived from 10 DAMMIF models, is shown in cyan spheres and has been optimally superimposed on the PLY structure. The normalised spatial discrepancy for the fit was 1.21.