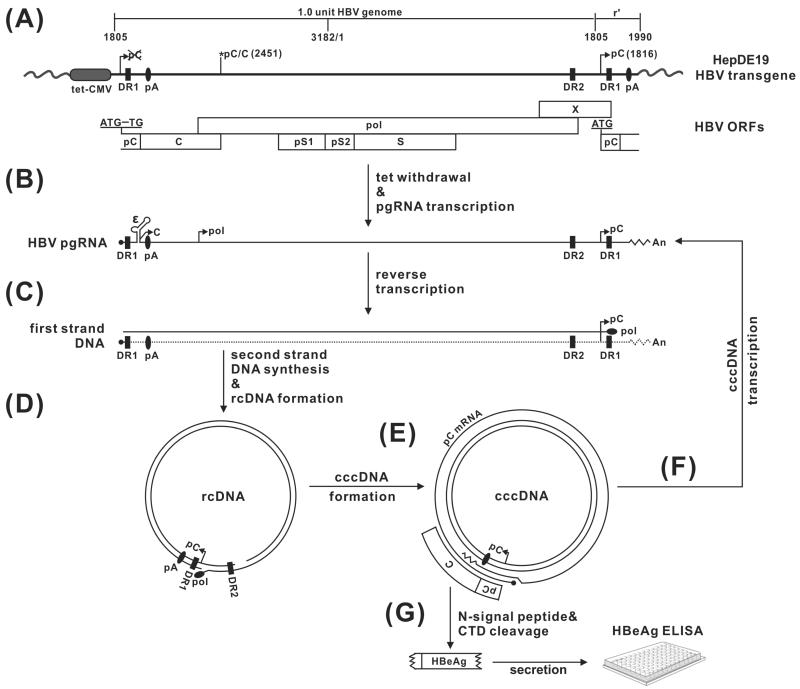
Fig 2. HBV nucleic acid metabolism in HepDE19 cells, with the rationale for design of a cccDNA-dependent HBeAg assay.
(A) The transgene in HepDE19 cells contains a 1.1 overlength HBV genome (genotype D, subtype ayw), starting from nt 1805, under the control of a tet-CMV promoter (tet-off), in which the start codon (AUG) of the precore (pC) ORF has been mutated to UG at the 5’ end of HBV DNA, with the second one at the 3’ redundancy (r’) left unchanged. C, pol, pS1, pS2, S and X represent ORF for core, polymerase, preS1, preS2 and S domain of HBV surface antigen, and X protein, respectively. DR represents the identical direct repeat sequences 1 and 2. pA is polyadenylation site. (B) Upon tetracycline withdrawal, HBV pgRNA is transcribed and the viral core and polymerase are produced, resulting in pgRNA encapsidation, (C) reverse transcription of pgRNA to minus-strand DNA, and (D) sequential plus-strand DNA synthesis and circularization into rcDNA. (E) rcDNA is converted to the cccDNA template, in which the precore ORF is restored, giving rise to authentic precore mRNA, and (F) pgRNA. (G) cccDNA-derived precore mRNA is translated into the precore protein, which is further processed into secreted HBeAg through proteolytic cleavage of the N-terminal signal peptide and C-terminal domain (CTD). The secreted HBeAg in the supernatant of HepDE19 cells can be detected by ELISA assay and serves as a surrogate marker for intracellular cccDNA.