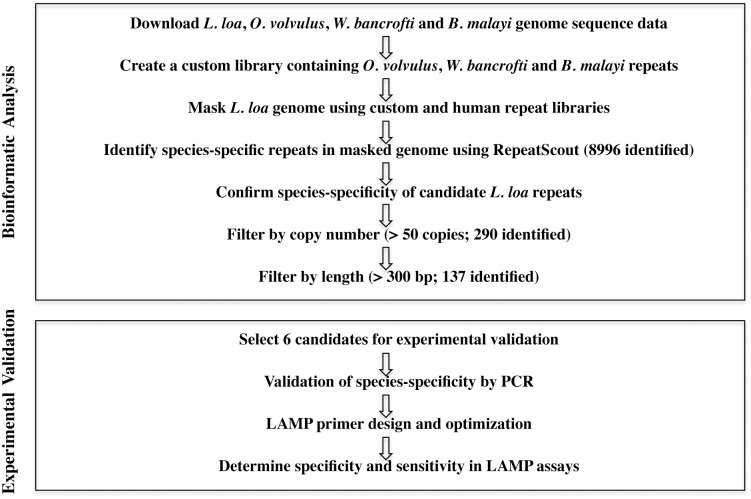
Fig 1. Bioinformatic filtering pipeline to identify L. loa-specific repeat families.
Genome sequences were downloaded from the Filarial Worms sequencing project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/). The L. loa genome was masked using RepeatMasker then mined for L. loa-specific repeats using RepeatScout. The resulting consensus repeat sequences that consist of 51 or more members and that are 300 bp or more in length were selected for further evaluation.