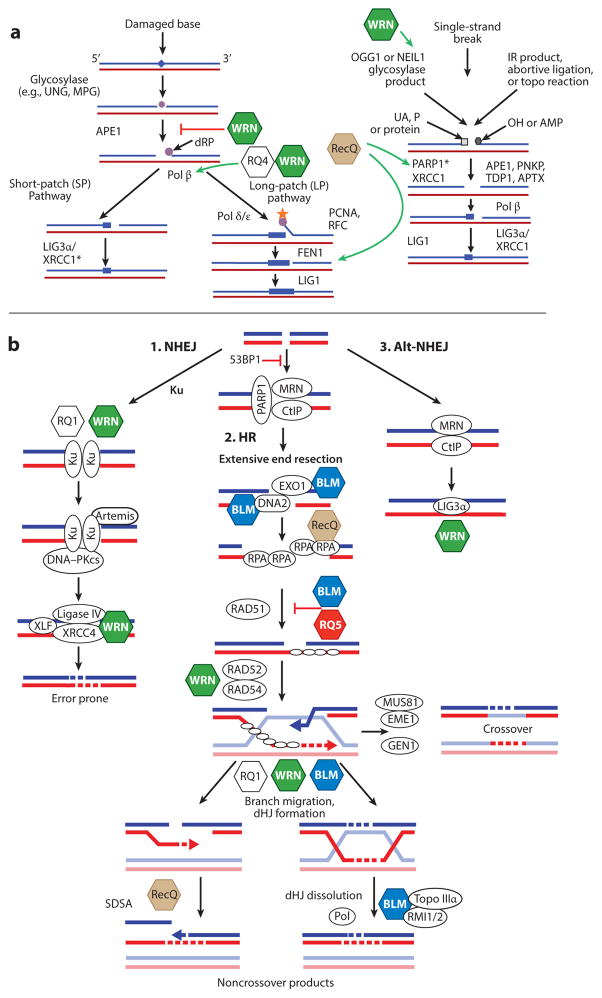
Figure 3.
RecQ helicases in base excision repair (BER) and double-strand break repair (DSBR). (a) Summary of the BER pathway. Interactions between RecQ helicases and BER proteins are depicted by a green arrow (for activation) or a red bar (for inhibition). Asterisks after PARP1 and XRCC1 indicate that these genes are specifically downregulated by RECQL5 loss. The brown RecQ hexagon indicates that multiple RecQs interact with these proteins. (b) Summary of DSBR pathways. (i ) Nonhomologous end joining (NHEJ). (ii ) Homologous recombination (HR). (iii ) Alternative (Alt)-NHEJ. The involvement of RecQ helicases in DSBR is depicted. Each RecQ helicase is represented as a colored hexagon. The colored hexagons are consistent with the coloring in Figure 2. Other key proteins are shown as white ovals. DNA molecules are shown as blue/red or light blue/red duplexes. Dashed lines denote nascent DNA synthesis. Abbreviations: dHJ, double Holliday junction; dRP, deoxyribose phosphate; IR, ionizing radiation; MPG, methylpurine DNA glycosylase; P, 3′-phosphate; PCNA, proliferating cell nuclear antigen; Pol, DNA polymerase; RFC, replication factor C; SDSA, synthesis-dependent strand annealing; Topo, topoisomerase; UA, 3′-α,β unsaturated aldehyde; UNG, uracil DNA glycosylase.