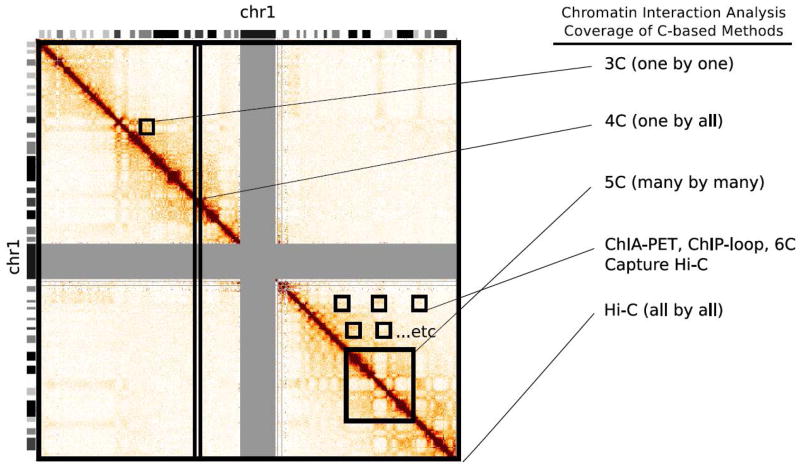
Figure 2.
An example Hi-C heatmap depicting the types of data generated by 3C or 3C-based methods. 3C queries interactions on a one-by-one basis; 4C identifies interactions between a single region and the rest of the genome; 5C generates a high-resolution interaction matrix of a large genomic region; Capture Hi-C, ChIA-PET and other ChIP-based methods characterize chromatin-looping interactions over several different loci; and finally Hi-C queries genome-wide chromatin interactions at low or high resolution.