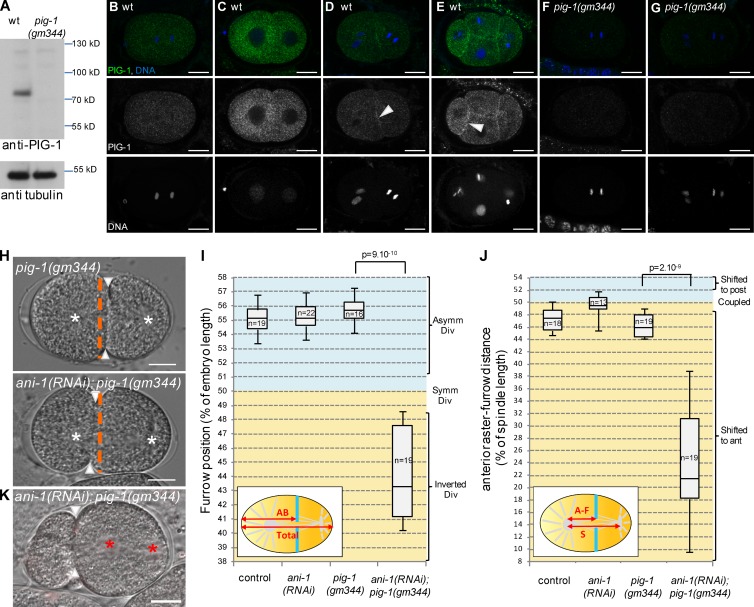
Figure 5.
Loss of PIG-1 and ANI-1 leads to strong furrow mispositioning. (A–G) PIG-1 is expressed in embryos. Western blot (A) shows that PIG-1 is expressed in wild-type but not pig-1(gm344) embryonic extracts. Tubulin is used as a loading control. Immunostainings (B–G) show that PIG-1 is expressed in early wild-type (B–E) but not pig-1(gm344) embryos (F and G). PIG-1 is localized in the cytoplasm in one-cell (B) and early two-cell (C) embryos, it is also localized at the cortex between adjacent cells in late two-cell (D) and four-cell embryos (E). See also Video 5. (H–J) ani-1(RNAi); pig-1(gm344) embryos show strong furrow mispositioning. DIC images (H) and quantification of furrow position (I) and furrow/spindle coupling (J) in dividing one-cell embryos of the indicated genotypes. In J, all strains also express an α-tub::YFP transgene. Furrow position was measured when the furrow reached its most anterior position. (H) Orange dashed lines correspond to the embryo center, arrowheads to furrow position, and white asterisks to spindle asters. See also Fig. S3. (K) Strong furrow mispositioning can lead to DNA segregation defects in ani-1(RNAi); pig-1(gm344) embryos. DIC pictures of an ani-1(RNAi); pig-1(gm344) embryo in which all DNA is inherited by the posterior cell. This embryo also expressed an mCherry::HIS-58 transgene to monitor DNA position (red asterisks). Arrowheads indicate furrow position. 12 out of 28 embryos showed this DNA segregation phenotype. Bars, 10 µm. P-values from Student’s t test.