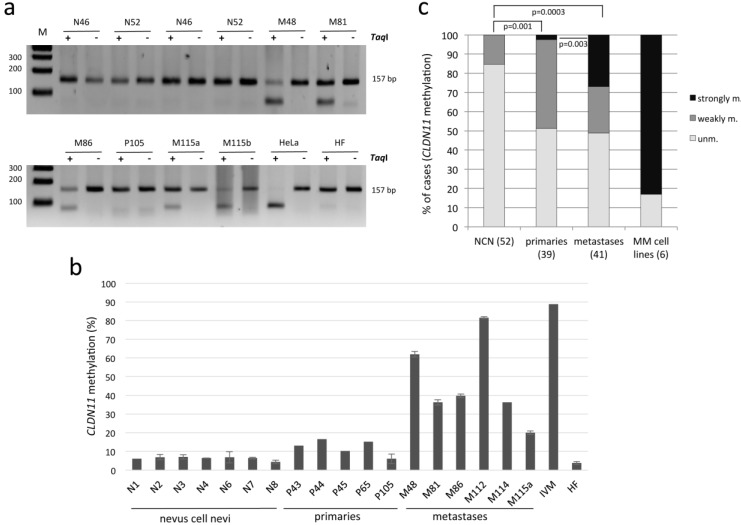
Figure 3.
Methylation of CLDN11 in nevus cell nevi (N), primary malignant melanoma (P) and melanoma metastases (M). (a) Methylation of CLDN11 was analyzed by COBRA. PCR products (157 bp) from bisulfite-treated DNA were digested with TaqI (+) or mock digested (−) and resolved on 2% gel. DNA from HeLa and fibroblast (HF) was utilized as methylated and unmethylated control. Sizes of a 100 bp ladder (M) are marked; (b) Methylation analysis of CLDN11 by bisulfite pyrosequencing. 11 CpGs within PCR products generated from nevus cell nevi (N), primary MM (P/primaries) and MM metastases (M) and in vitro methylated DNA (IVM) were analyzed in technical replicates. M48 is a lymph node metastasis and all other metastases originated from skin; (c) Comparison between methylation of MM (primaries, metastases) and NCN. Bar charts indicate percentage of cases with unmethylated (unm. <10%), weakly methylated (10% to 20%) and strongly methylated (>20%) CLDN11. Significance was calculated with the two-tailed Fisher exact probability test.