Fig. 4.
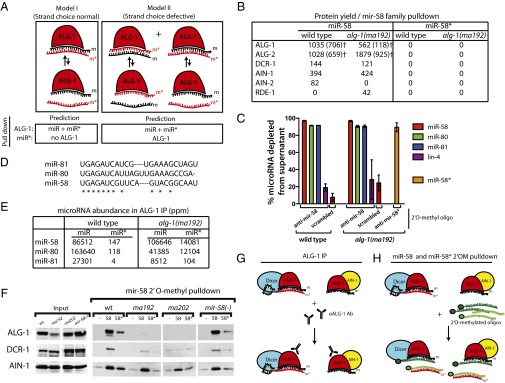
2′O-methyl oligonucleotide pull-downs from wild-type and alg-1(anti) mutants suggest that miR-58* accumulation in ALG-1 is a result of failure of ALG-1(anti) to release it from the duplex. (A) Two models for miR* microRNA accumulation in alg-1(anti) mutant animals. Model I: ALG-1(anti) complexes retain both microRNA strands primarily in a bound duplex. Model II: ALG-1(anti) complexes contain single strands of miR and miR* microRNAs because of a loss in the ability of the protein to differentiate between the two strands. Predictions of the outcome for IP and pull-down experiments are shown directly below each model. (B) Protein yield of miRISC components and other small RNA related factors from mir-58 family microRNA pull-down as determined by MudPIT proteomics and normalized to the amount of RNA depleted by the 2′O-methyl pull-down. †ALG-1 and ALG-2 protein yield as determined based on normalized spectral abundance factor (NSAF)unique (SI Materials and Methods). (C) Efficiency of the miR-58 and miR-58* strand pull-downs presented as percent microRNA depleted from the supernatant in samples shown in B. (D) Sequence alignments between members of the mir-58 family of microRNAs. (E) miR-58 family microRNA and miR* abundances in wild-type ALG-1 and ALG-1(anti) IP. (F) Western blot analysis of mir-58 microRNA family pull-downs from extracts of alg-1(anti) and wild-type animals showing association of ALG-1, DCR-1, and AIN-1 with mir-58 family microRNAs (-, scrambled oligo control; 58, oligo against miR-58; 58*, oligo against miR-58*). (G and H) Summary model for the miR-58* strand and related proteins accumulation as observed in alg-1(anti) mutants by (G) ALG-1(anti) IP and (H) miR-58 and miR-58* 2′O-methyl pull-downs (m, miR; m*, miR*).
