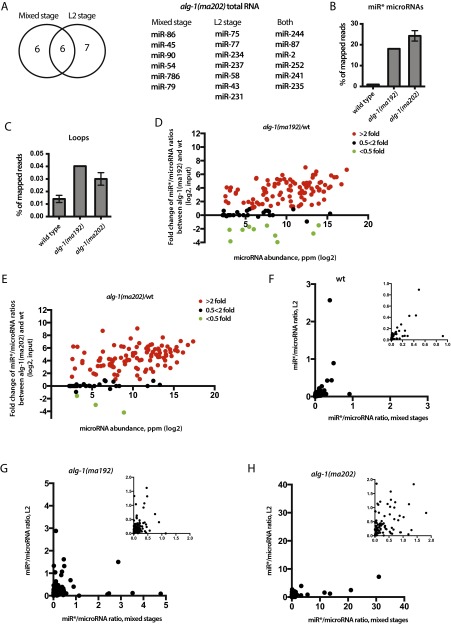
Fig. S4.
alg-1(anti) second larval (L2)-stage animals accumulate miR* strands. (A) miRNAs that showed asymmetry (miR*/miR >1) in the total RNA of alg-1(ma202) mixed-stage populations overlapped with miRNAs that showed asymmetry in the total RNA of alg-1(ma202) L2 stage animals. (B and C) Deep-sequencing analysis of cDNA libraries from small RNA populations shows an increase in miR* (B) and loop (C) levels in alg-1(ma192) and alg-1(ma202) animals compared with wild-type. (D and E) Scatterplots showing fold-change in individual miR*/miR ratios between alg-1(anti) and wild-type total RNA. Increased miR*/miR ratio is seen in alg-1(ma192) (D) and alg-1(ma202) (E) mutants and is independent of microRNA abundance. Red dots represent microRNAs with an increased miR*/miR ratio in the alg-1 mutant over wild-type of at least twofold; green dots represent miRNAs with a decreased miR*/miR ratio in the alg-1 mutant compared with wild-type of at least twofold. Black dots represent no change in miR*/miR ratio in the alg-1 mutants compared with wild-type. (F–H) Scatterplots showing miR*/miR ratios observed in mixed-stage vs. L2-staged populations in wild-type (F), alg-1(ma192) (G), and alg-1(ma202) (H) animals. F–H, Insets show zoomed-in graphs.