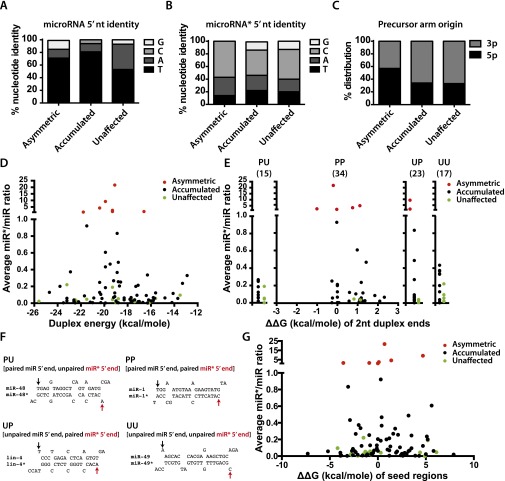
Fig. S7.
microRNA feature analysis of microRNAs with a miR*/miR ratio of >1 in ALG-1(ma202) IP (termed asymmetric microRNAs) (class I in Table 1); classes II and III, microRNAs that showed an increase of miR*/microRNA ratio in ALG-1(ma202) compared with wild-type ALG-1 (accumulated) (Table 1); and the microRNAs whose miR*/microRNA ratio was not affected (termed “unaffected”) (class IV in Table 1). (A) The identity of the 5′ nucleotide of the guide microRNA. (B) The identity of the 5′ nucleotide of the miR* microRNA. (C) Precursor arm origin. (D) Duplex energies comparison between the asymmetric (class I in Table 1), accumulated (classes II and III in Table 1), and unaffected (class IV in Table 1) microRNA populations. (E) Differences in the end-pairing and duplex end energies (ΔΔG) of the three microRNA populations. PP, paired 5′ nucleotide of the microRNA, paired 5′ nucleotide of the miR*; PU, paired 5′ nucleotide of the microRNA, unpaired 5′ nucleotide of the miR*; UP, unpaired 5′ nucleotide, paired 5′ nucleotide of the miR*. Number in parenthesis represents the total number of microRNAs within each group. ΔΔG(kcal/mole) of two nucleotides at each end of duplex is plotted as a function of miR*/microRNA ratio for the microRNA duplexes that had paired nucleotides at each duplex end. (F) MicroRNA duplex examples of the four groups (PU, PP, UP, UU) shown in E. (G) ΔΔG(kcal/mole) of seed energies (ΔG of miR seed minus ΔG of miR* seed) is plotted as a function of miR*/miR ratio for the microRNA duplexes.