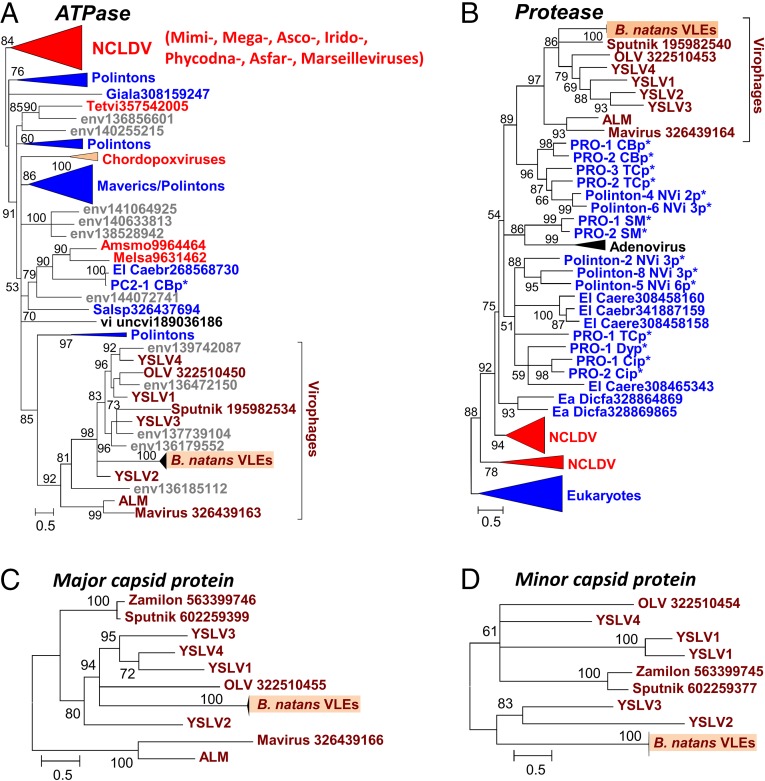
Fig. 3.
Maximum-likelihood phylogenetic trees of conserved virophage proteins. (A) Packaging ATPase. (B) Maturation protease. (C) Major capsid protein. (D) Minor capsid protein. Branches with bootstrap support (expected-likelihood weights) less than 50% were collapsed. Sequences marked with an asterisk were taken from Repbase (38). For other sequences, the species name abbreviation and GenBank accession number are indicated; env, marine metagenome. Species: Amsmo, Amsacta moorei entomopoxvirus “L”; Caebr, Caenorhabditis brenneri; Caere, Caenorhabditis remanei; Dicfa, Dictyostelium fasciculatum; Giala, Giardia lamblia; Melsa, Melanoplus sanguinipes entomopoxvirus; Salsp, Salpingoeca rosetta; Tetvi, Tetraselmis viridis virus; uncvi, uncultured virus. Taxa: Ea, Amoebozoa; El, Opisthokonta; u2, Entomopoxvirinae. Dark red, virophages; blue, (predicted) polintons and related elements; light red, NCLDV; gray, unassigned environmental sequences. The numbers of validated amino acid positions in cleaned alignments are 210 (A), 166 (B), 548 (C), and 408 (D).