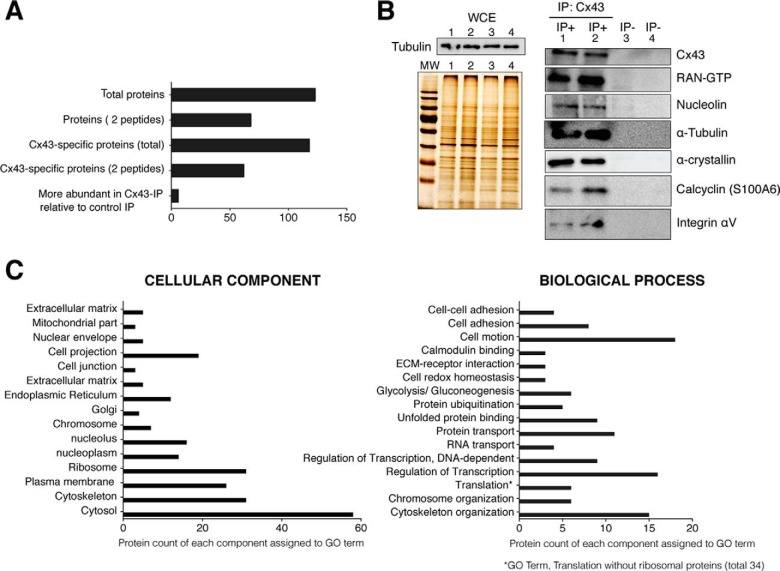
Fig. 1.
Analysis of Cx43-associated proteins identified using mass-spectrometry. Primary chondrocytes in culture were lysed in the presence of the nonionic surfactants Igepal and Tripton X-100 using a near-physiological-salt concentration of 150 mm NaCl to pull-down the Cx43 (nonjunctional and junctional Cx43). Immunoprecipitated proteins using an antibody that specifically recognized the C-terminal tail of Cx43 were identified using MS. Four different samples of chondrocytes from four healthy donors were analyzed. Overall, proteins were identified with 95% peptide confidence (minimum value 1.3 of unused cutoff) A, Summary of the number of identified proteins. Proteins identified as specific to the Cx43-IP but absent from the control IP (no antibody). The “more abundant proteins” in Cx43-IP relative to control IP correspond to the number of proteins identified as most likely more abundant in the Cx43-IP than in the control-IP (peptide difference ≥2). See supplemental Tables S1 and S2 for a complete list of proteins identified in the Cx43-IP and control IP. B, Whole-cell extracts (WCE) and proteins immunoprecipitated with (IP+) or without (IP−) an antiCTD-Cx43 antibody were analyzed by silver staining and Western blotting. The protein content of the WCE (input, without antibody) (images shown on the left) for each experiment (IP) (images shown on the right) was previously analyzed by silver staining and Western blotting (Tubulin) (image shown on the top-left). Lanes represent 0.5% of the total WCE used for each IP. The numbers (1–4) represent the WCE utilized for the corresponding experiment (IP). For the images shown on the right, lanes represent the 25% of each IP C, Gene Ontology clustering of the protein set. GO analysis (27, 28) for identified proteins reveals a strong enrichment for GO terms associated with cytoskeleton, nuclear localization, translation processes, cell adhesion, and motion and carbohydrate metabolism. See supplemental Tables S1 and S2 for a complete list of the identified proteins.