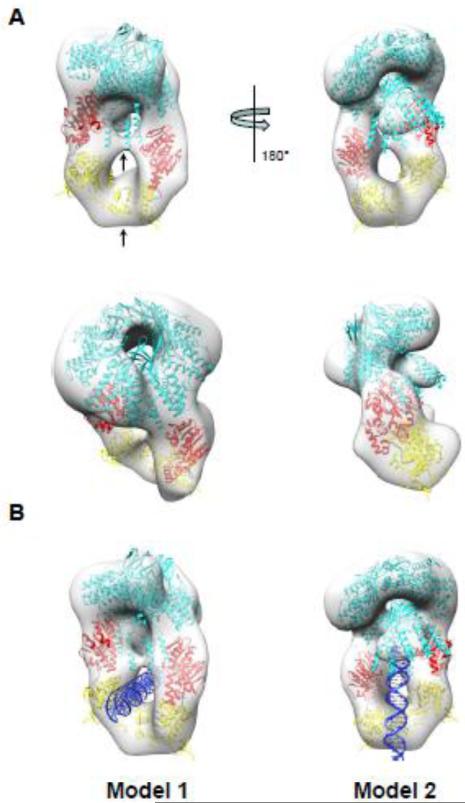
Figure 6. Pseudo-atomic model of S:L-terminase holoenzyme.
(A) Various views of the 3-D reconstruction in Fig. 5B colored as a transparent gray surface with docked models of one nonameric S-terminase (cyan) and two L-terminase subunits with ATPase and nuclease domains colored in red and yellow, respectively. Each L-terminase has an ATPase domain (red) and a nuclease domain (yellow) separated by a short flexible linker (black). Models were rigid-body refined into density using the “Fitmap” function in Chimera [50]. (B) Models of S:L-terminase binding to dsDNA. In model 1, dsDNA passes through the lower hole between the L-terminase domains. In model 2, the dsDNA is vertically positioned between the two L-terminase domains and abuts the S-terminase C-terminal helices. In this conformation, dsDNA could fit in the S-terminase central channel.