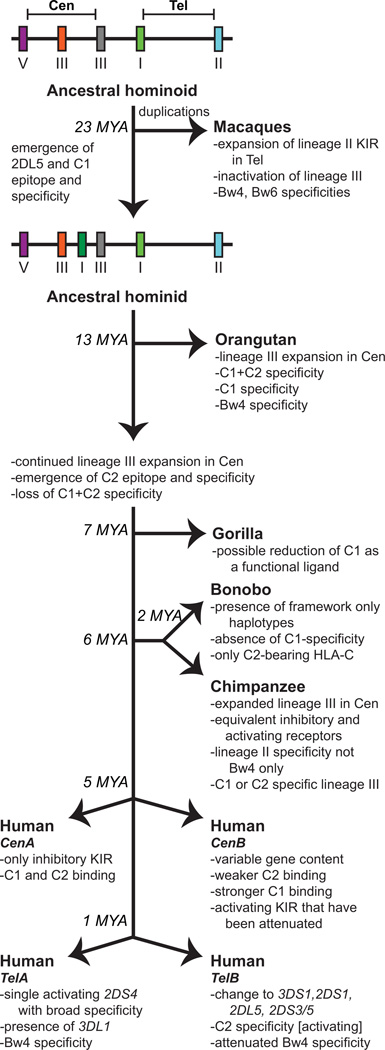
Figure 13. Summary of the co-evolution of MHC class I and KIR in the catarrhine primates.
This schematic diagram shows the distinguishing features of each of the KIR regions in the species shown. Approximate speciation times are indicated at the branch points (MYA, million years ago). Prior to the divergence of Old World monkeys (macaques) and apes, the KIR region minimally contained the framework genes and a lineage III gene as is shown at the top of the figure in the ancestral catarrhine KIR haplotype. Following this speciation event, the macaques expanded the number of lineage II KIR genes through duplication and recombination. In the hominid branch, KIR2DL5 emerged to form the ancestral hominid KIR haplotype. Specific adaptations that occurred subsequently in particular ape species are indicated. For humans, the emergence of distinctive CenA and CenB segments occurred ~5 MYA followed by the formation of distinctive TelA and TelB ~1 MYA. The distinguishing characteristics of these segments are described. Human KIR A haplotypes are defined by a fixed gene content and comprise CenA/TelA. KIR B haplotypes contain at least one of the characteristic B haplotype genes and are much more variable in KIR gene content than the KIR A haplotypes.