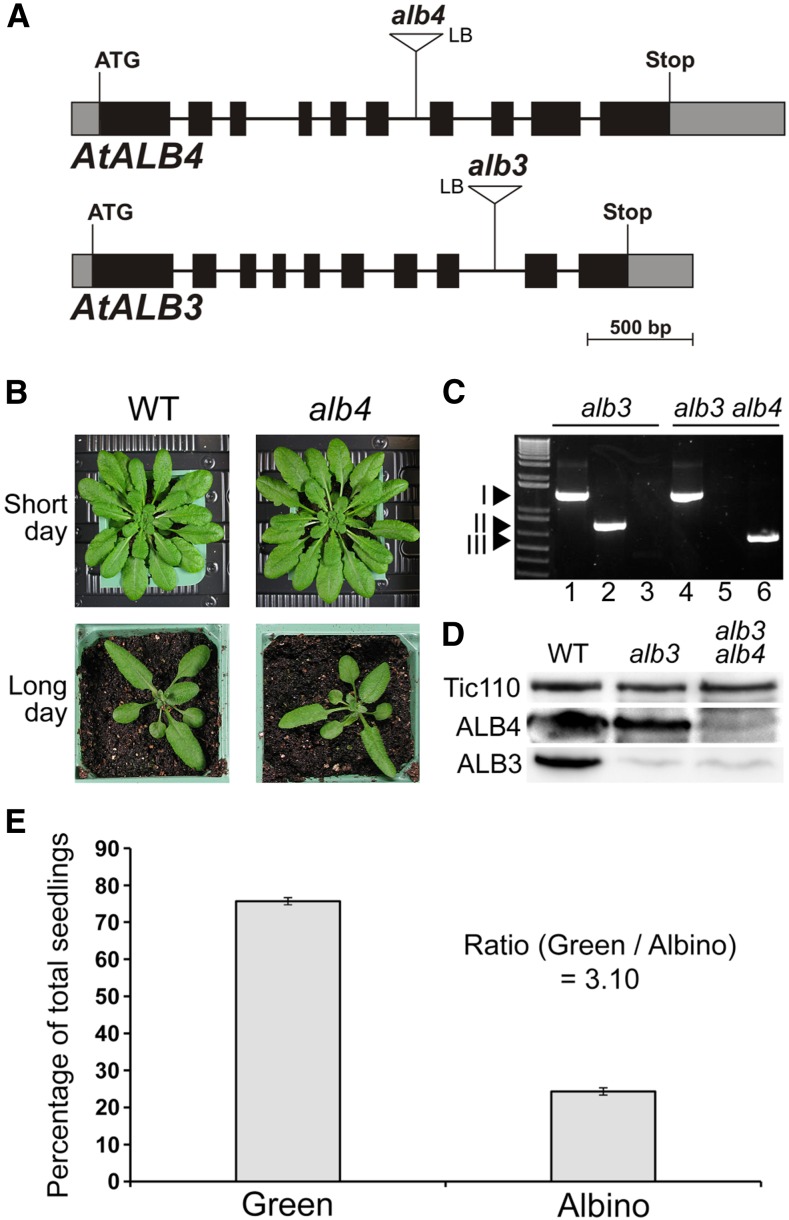
Figure 1.
Basic characteristics and genetic analysis of the Arabidopsis alb4 and alb3 mutants. A, Gene diagrams of Arabidopsis ALB4 and ALB3 with the T-DNA insertions of the alb4 and alb3 mutant lines shown. Protein-coding exons are represented by black boxes, untranslated regions by gray boxes, and introns by thin lines between the boxes. T-DNA insertion sites are indicated precisely, but insertion sizes are not to scale. ATG, Translation initiation codon; Stop, translation termination codon; LB, T-DNA left border. B, Wild-type (WT) and alb4 mutant plants (Salk_136199) were directly grown on soil under short-day (8-h-light/16-h-dark) conditions or long-day (16-h-light/8-h-dark) conditions. Pictures were taken after 7 weeks (short day) and 4 weeks (long day). C, Genotype analysis by genomic PCR. Heterozygous alb3 mutants (GK_293B08) and alb3/+ alb4/alb4 plants were visibly indistinguishable from the wild type, but both contained the alb3 T-DNA insertion (I; lanes 1 and 4); alb3/+ alb4/alb4 plants additionally contained the homozygous alb4 T-DNA insertion (III; lanes 3 and 6) but not the wild-type ALB4 allele (II; lanes 2 and 5). The ladder (lane 0, left side) includes standards of the following sizes, starting at the bottom: 0.3, 0.4, 0.5, 0.65, 0.85, 1.0, 1.65, and 2.0 kb. D, Immunoblot analysis of total protein extracts from 3-week-old albino plants. The alb3 alb4 double-mutant seedlings contained considerably less ALB4 protein than alb3 single mutants. The translocon at the inner chloroplast envelope membrane component of 110 kD (Tic110) has been used as a loading control. E, Segregation analysis of the progeny of three individual alb3/+ alb4/alb4 (heterozygous/homozygous) plants. Values are means of the percentages of green and albino progeny from each of the three parent plants. The error bars denote sd.