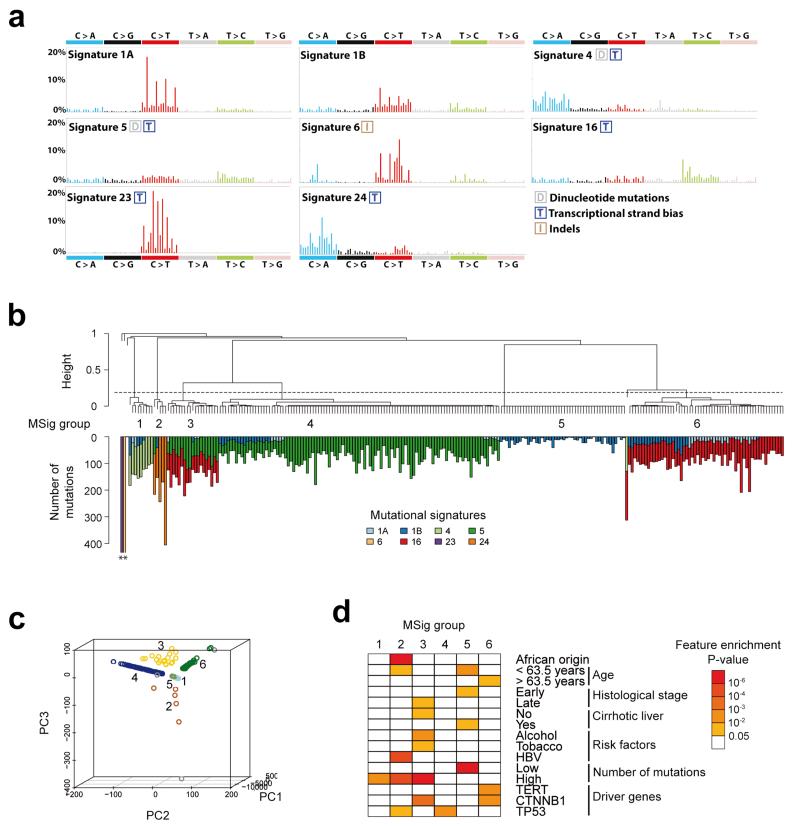
Figure 1.
Consensus signatures of mutational processes in hepatocellular carcinoma. (a) Patterns of the signatures of the mutational processes operative in 243 liver exomes. Signatures 23 and 24 were identified using de novo WTSI mutational signatures framework, while the presence of Signatures 1A, 1B, 4, 5, 6, and 16 were identified via re-introduction of consensus mutational signatures previously identified in liver cancer by a pan-cancer analysis14. Each signature is displayed according to the 96 substitution classification defined by the substitution class and sequence context immediately 3′ and 5′ to the mutated base. Signatures 1A, 1B, 4, 5, 6 and 16 match signatures previously described in a pan-cancer study14 and the plotted patterns correspond to the updated consensus signatures. Signatures 23 and 24 are new.(b) Unsupervised hierarchical clustering of 243 liver tumors based on the intensity of signatures operative in each sample. Tumors were classified into 6 mutational signature (MSig) groups and 4 singletons. The number of mutations attributed to each signature in each tumor is represented by color bars below the dendogram. *Tumors harboring more than 500 mutations. (c) Principal component analysis of mutational signature intensities in 243 liver tumors. Tumor samples are plotted in three dimensions using their projections onto the first three principal components (PC). MSig group membership is represented by a color code and labels. (d) Clinical and molecular features associated with each MSig group. Associations were assessed using Chi-square tests for categorical variables and ANOVA for quantitative features.