Fig. 2.
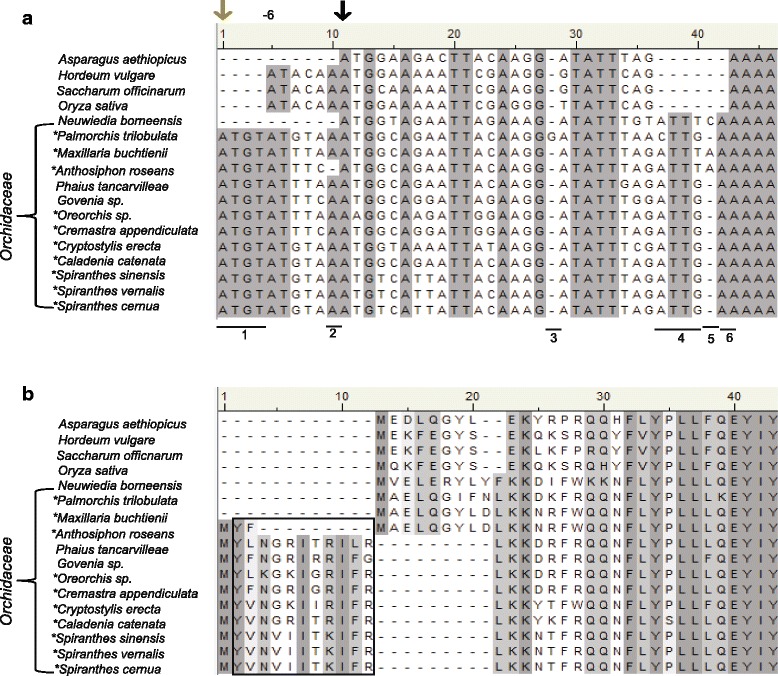
Sample alignments showing the alternative initiation codon (aic) and subsequent MatK translation products. (a) Nucleotide alignment starting at the aic of the same sample orchid species and non-orchid monocot species (Asparagus aethiopicus, Hordeum vulgare, Saccharum officinarum and Oryza sativa) as in Figure 1. Asterisks indicate species noted in GenBank to contain matK as a pseudogene. Gray arrow shows hypothesized out-of-frame alternative initiation codon. Black arrow indicates consensus initiation codon (cic) used by other monocots and angiosperms for matK expression. The ‘-6’ indicates the position of the upstream initiation codon in-frame with the cic. Underlined sections are indels that realign the matK reading frame for translation using the consensus vs. the alternative initiation codon. Five prime untranslated region of matK from Hordeum vulgare, Saccharum officinarum and Oryza sativa shown for contrast in sequence between non-orchid monocots and orchids in this region. (b) Translated MatK amino acid sequence using the aic for species previously containing premature stop codons when translated with the cic (Phaius tancarvilleae, Govenia sp., Oreorchis sp., Cremastra appendiculata, Cryptostylis erecta, Caladenia catenata, Spiranthes sinensis, S. vernalis and S. cernua). Asterisks in translated amino acid sequence indicates stop codons in the MatK reading frame. Large gaps in N-terminus of MatK amino acid sequence are indicative of the 11 amino acids different between translations using the aic vs the cic, not true indels. The 11 amino acids that differ due to translation initiation codon are boxed for clarity. Gray shadowing indicates highly conserved sequence among all taxa in the alignment, lighter gray indicates less conserved sequence, white background indicates a lack of conserved sequence
