Fig. 3.
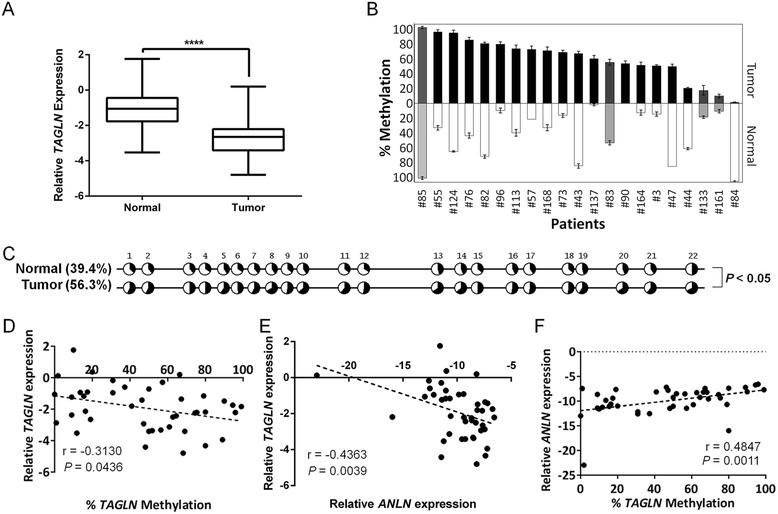
Expression and promoter methylation analyses of TAGLN gene in normal matched tumor tissues. a Box-plot analysis showing strong downregulation in average TAGLN expression in tumor tissues compared to matched normal tissues. Log2 expression levels relative to geometric means of ACTB and SDHA reference genes were determined by qRT-PCR. ****P < 0.0001, Wilcoxon signed rank test. b Comparative bar charts of bisulfite sequencing based percent methylation scores of matched tumor and normal pairs showing significance hypermethylation in majority of tumors. Methylation percentages represent the average of five clones for each tissue. Black-white bar pairs represent significant (P < 0.05) differences between tumor and normal tissues (Wilcoxon signed rank test). Error bars represent SEM values. c Overall average methylation levels based on QUMA were significantly higher in breast tumors compared to normal tissues; Wilcoxon signed rank test. Methylated DNA ratio; black area and unmethylated; white area in the pie chart. d–f Scatter plots show correlations between relative TAGLN expression levels and methylation status of TAGLN promoter in tumor and normal tissues; Spearman r = −0.3130, P = 0.0436 d; relative TAGLN and ANLN expressions; Spearman r = −0.4363; P = 0.0039 e; and relative ANLN expression and TAGLN methylation; Spearman r = 0.4847, P = 0.0011 f. All expression values are log2. The correlations were robust to outliers
