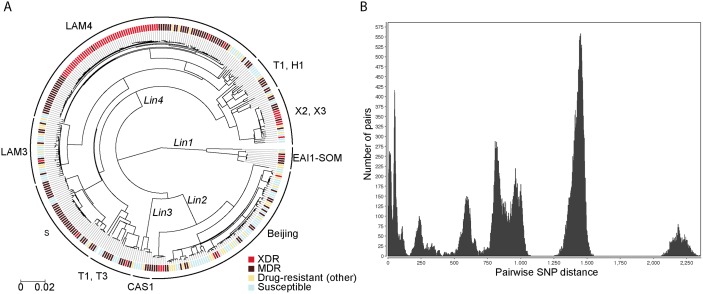
Fig 1. Diverse strains contribute to drug resistance in KwaZulu-Natal.
(A) Midpoint rooted maximum-likelihood phylogeny of 340 M. tuberculosis isolates. Four of the seven known M. tuberculosis lineages were identified: CAS (Lin1), Beijing (Lin2), EAI (Lin 3), and Euro-American (Lin4). Digital spoligotyping identified 17 unique spoligotypes in the dataset; spoligotypes are shown on this figure if they are represented by three or more strains. Corresponding spoligotypes and phenotypes are reported for all strains in S4 Table. Phenotypic XDR, MDR, poly- and monodrug resistance (labeled “Drug-resistant other”), and pansusceptible strains are indicated by colored tick marks at the tip of each leaf node. (B) Histogram of pairwise SNP distances between strains. The number of pairs within each SNP distance range is plotted. The peaks correspond to the distance between major lineages. The peak at the far left of the figure corresponds to the distance between pairs of strains within a clone.