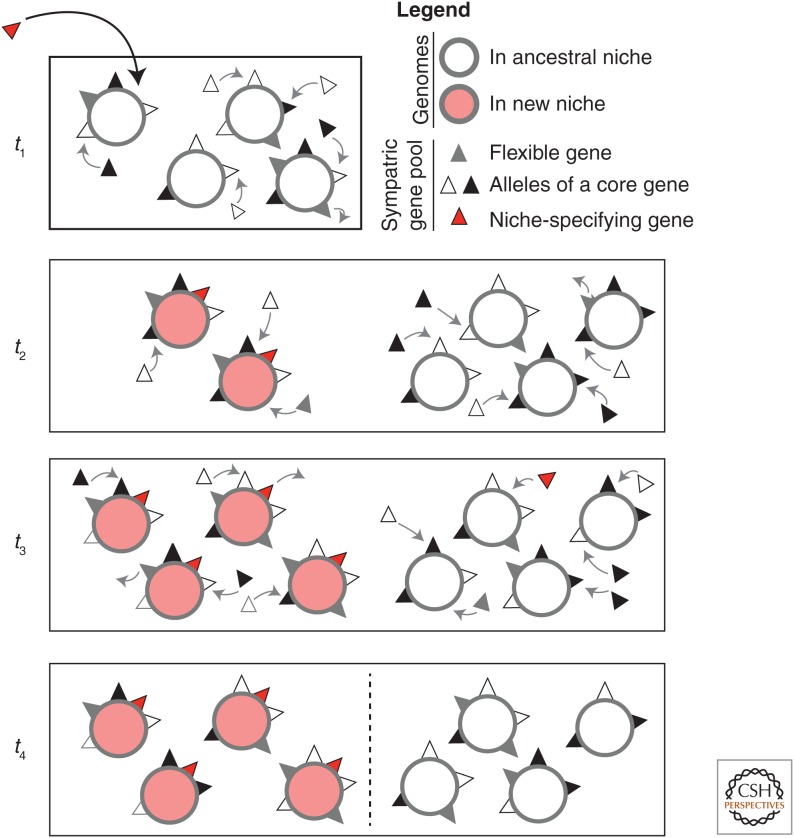
Figure 3.
Ecological differentiation via a gene-specific sweep. This illustration follows the basic steps of the symsim model (Friedman et al. 2013). At the first time point (t1), a niche-specifying gene (red triangle) arrives into a homogeneously recombining population occupying a single niche. Between t1 and t2 (as between all time points), recombination (r) occurs at random from a sympatric pool (one to two events per genome, illustrated as arrows), then genomes reproduce clonally, and are culled to a carrying capacity of four genomes per niche. Because the niche-specifying gene confers a selective advantage(s) in the new niche, genomes that contain it grow exponentially until the carrying capacity is reached at t3. Other genomes are culled at random, because the rest of the gene pool is neutral to fitness. By t4, the gene-specific sweep is complete. The niche-specifying gene is in perfect association with the new niche, but all other genes are randomly distributed across niches. At this point, barriers to recombination between niches (dashed line) may or may not emerge. (Note that recombination events at t4 are not shown for purposes of clarity, but this does not mean they do not occur.)