Fig. 5.
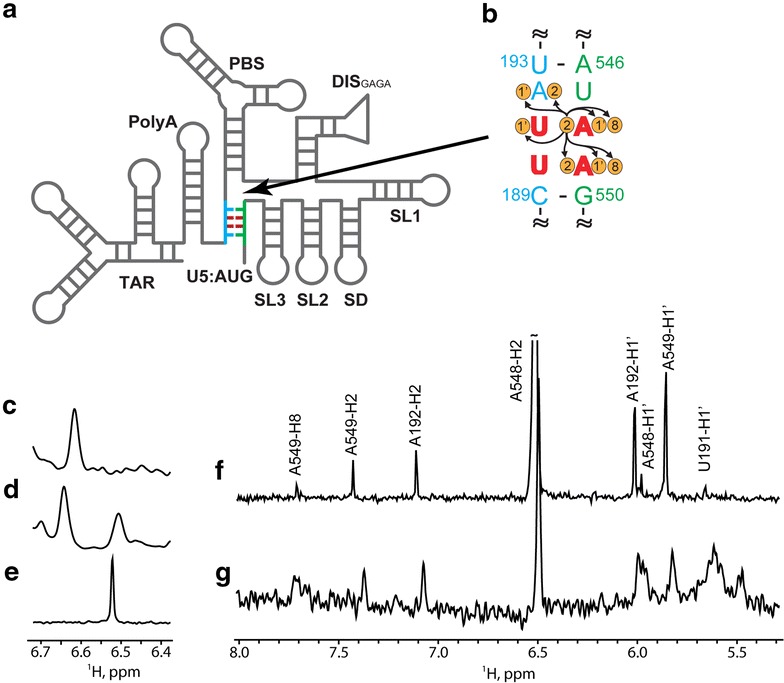
NMR detection for HIV-2ROD U5:AUG formation. a Schematic representation of the dimeric HIV-2 5′-leader showing DIS exposure and long range base pairing between the U5 (cyan) and AUG (green) elements. b Portion of the lr-AID modified U5:AUG helix showing C-G to U-A substitutions in red. c–e Portions of the 1D 1H NMR spectra obtained for HIV-2ROD 5′-L, lr-AID substituted HIV-2ROD 5′-LGAGA, and a control oligo-RNA containing the lr-AID substituted U5:AUG stem, respectively. Signals at ~6.51 ppm are due to the A548-H2 (lr-AID) signal. f F2 slice from the 2D NOESY spectrum obtained for the control lr-AID U5:AUG oligonucleotide. Signal assignments were made by standard connectivity methods [57]. g 1D saturation-difference NOE spectrum obtained for lr-AID substituted HIV-2ROD 5′-LGAGA. Except for expected intensity differences (due to differences in rotational correlation time and NOE mixing periods employed), the spectral features are similar to those of the control RNA, indicating that both RNAs adopt the predicted U5:AUG helix
