Figure 1. Genomics-based identification of neoepitopes.
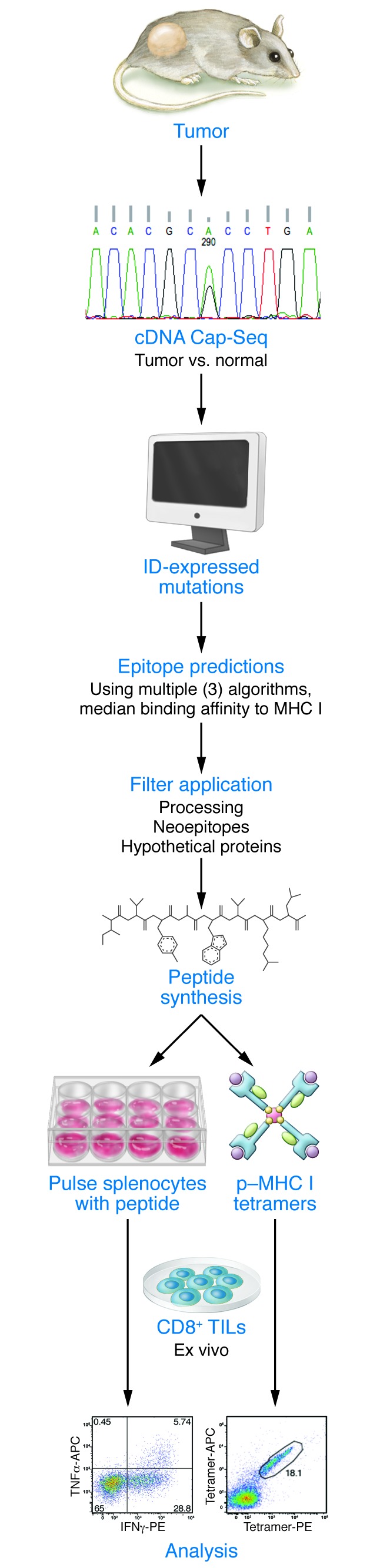
Tumor cells and normal tissue were subjected to cDNA Cap-Seq to identify expressed, nonsynonymous somatic mutations. Corresponding mutant epitopes were then analyzed in silico for MHCI binding. Filters were applied for antigen processing, neoepitopes, and deprioritization of hypothetical proteins. Peptides corresponding to predicted epitopes were then synthesized and used to identify mutant neoantigen–specific T cells in freshly explanted TIL using MHC multimer–based screens or cytokine induction by peptide stimulation.
