Abstract
Most of what we currently know about how neural circuits work we owe to methods based on the electrical or optical recording of neural activity. This is changing dramatically. First, the advent of optogenetic techinques has enabled precise manipulation of the activity of specific neurons. Second, the development of super-resolution methods for obtaining detailed maps of synaptic connectivity has paved the way for uncovering the connectomes of entire brains or brain regions. We describe a third and complementary new strategy for investigating and manipulating neural circuits: the artificial insertion of new synapses into existing neural circuits using genetic engineering tools. We have successfully accomplished this in C. elegans. Thus, In addition to being the first animal with an entirely mapped connectome, C. elegans is now also the first animal to have an editable connectome. Variations on this approach may be applicable in more complex nervous systems.
Keywords: connectome, connexin, electrical synapse, gap junction, innexin, synaptic engineering, synapse
Why Engineer Synaptic Connections?
Synapses are a fundamental building block of neural circuits. The pattern of synaptic connectivity directs the spatial and temporal flow of information through the circuit, determining its function and ultimately affecting behavior. For this reason a tremendous research effort is currently being made to obtain detailed connectomes, whole brain synaptic connectivity maps, of various organisms, including humans.1-3 This formidable endeavor follows the earlier, relatively more modest project of mapping the entire C. elegans connectome almost 3 decades ago,4-6 which has continually proven to be of enormous value.
Nevertheless, a functional understanding of neural circuits requires a functional analysis of the structure revealed by connectomics. Much information can be gained from recording activity patterns in identified circuits and from molecular characterization of individual mapped synapses.7 However, observation and mapping are not sufficient; in addition, an engineering approach, similar to that underlying synthetic biology, whereby individual biological components are artificially reassembled or controlled to determine the effect on system output,8-11 provides a critical test for functional importance. Indeed, optogenetic techniques to artificially manipulate neuronal activity at high spatio-temporal resolution have been transformative for neuroscience.12,13 In a similar manner, techniques to synthetically modify a neuron's connectivity14 could offer new opportunities for addressing fundamental questions regarding the relationship between synaptic connectivity and neural circuit function. For example, could several alternative patterns of synaptic connectivity implement similar functions? What changes in synaptic connections are sufficient to significantly alter behavior? And can we rationally design new kinds of behaviors or repair malfunctioning circuits by modifying synaptic connections artificially?
How to Insert a Synapse Into the Connectome?
Several techniques exist for manipulating synaptic transmission. Pharmaceutical and genetic silencing or activation of neurotransmitter release or reception mechanisms is the most traditional and widely used. However, these methods mostly target the overall transmission or reception properties of neurons or neuronal populations and thus affect the total neuronal output or input, so that the effective unit of manipulation is actually the neuron rather than a specific synapse (Fig. 1A).
Figure 1.
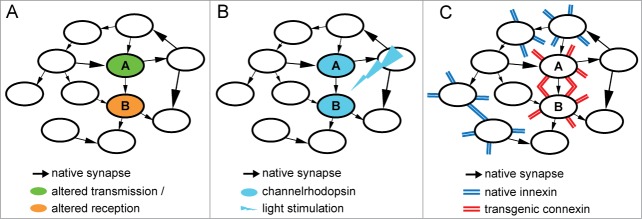
Strategies for manipulating a synaptic connection between 2 neurons, A and B. (A) Pharmaceutical or genetic silencing or over-activation of either presynaptic or postsynaptic components. (B) Optogentic induction of long-term potentiation or depression of the synaptic connection between 2 light-stimulated neurons. (C) Transgenic expression of vertebrate gap junction connexin proteins in invertebrate neurons.
Recently paired optogenetic stimulation of neurons has been used to target synaptic connections by inducing in existing synapses long-term potentiation (LTP) or depression (LTD), 2 forms of timing-dependent synaptic plasticity15,16 (Fig. 1B). Although effective, this method has several drawbacks. First, its indirect nature implies that it might induce diverse and unpredictable collateral modifications in other synapses and neurons. For example, it might induce plasticity of target neurons’ intrinsic excitability, altering ionic conductances,17,18 or it might affect synaptic connections other than the targeted ones through non-Hebbian (non coincidence-dependent) mechanisms.19,20 Second, the pairing protocols and their effects on LTP and LTD direction, magnitude and stability may vary considerably between specific synaptic partners and between preparations,21 requiring ad hoc solutions for each particular synaptic manipulation. Third, it relies on the ability to deliver light to the target neurons, which might be challenging.
Instead, we have devised a fundamentally different strategy, comprising the direct and specific insertion of new synapses into neural circuits using genetic engineering tools. We have successfully applied this method to C. elegans, and were thus able to edit its connectome.22 How is this done? Our goal was to introduce a new transgenic synapse between 2 neurons A and B. We reasoned that inserting a new chemical synapse might be difficult, since this should entail ectopic expression of many, possibly hundreds of constituent proteins on both the presynaptic and postsynaptic sides23,24 of the engineered connection. Moreover, this strategy might generate improperly assembled complexes or interfere with existing synaptic machinery. Consequently, we took advantage of the relative simplicity of electrical synapses.25 These are formed by the joining of 2 hemi-channels into a gap junction that can directly transfer electrical charge between 2 neurons. Each hemi-channel consists of as little as one gap junction protein type, belonging in invertebrates to the innexin family or in vertebrates to the connexin family.26 These 2 protein families are completely distinct in sequence, and yet they are strikingly similar in function. Importantly, although gap junctions may contain more than one type of connexin or innexin, attempts to induce hybrid connexin-innexin gap junctions have failed.27 Our strategy thus consisted of heterologously expressing a vertebrate connexin (we chose a brain ubiquitous mouse connexin called Cx3628) in adjacent C. elegans neurons using cell-specific promoters. Since connexins should not interact with endogenous innexins from other neighboring neurons, we expected a new gap junction to form exclusively between connexin-expressing neurons A and B (Fig. 1C). Indeed, Cx36 readily expressed in a variety of C. elegans neurons in a synapse-like punctate pattern.22 Calcium imaging experiments demonstrated the formation of new functional electrical synapses following simultaneous expression of Cx36 in the 2 neurons, but not when Cx36 was expressed only in one of the neurons.22
Examples of Synaptic Engineering Applications
Adding gap junctions to existing electrical synapses
The C. elegans response to nose touch is controlled by a circuit consisting of several sensory neurons, CEP, OLQ, FLP, that are each connected by electrical synapses to an interneuron, RIH. This hub-and-spoke circuit motif seems to be over-represented in the C. elegans connectome.5,29 We found that this nose touch circuit acts as a coincidence detector,30,31 displaying a substantial difference in circuit output when all sensory neurons are activated at the same time (Fig. 2A) compared to partial activation (Fig. 2B). Modeling work that we conducted suggested that the reduced output might stem from shunting of current through electrical synapses away from the output neuron, RIH, into the inactive sensory neurons31 (e.g. CEP in Figure 2B; arrow from RIH to CEP). The model further predicted that if the electrical coupling between RIH and the silent sensory neuron, CEP, were to be enhanced then the shunting inhibition would be stronger and the RIH output would become even smaller (Fig. 2C). We were able to test this hypothesis by inserting electrical synapses composed of Cx36 between RIH and CEP and thus increasing the electrical coupling between these neurons (Fig. 2C, enlarged arrow from RIH to CEP). As predicted, the RIH output became significantly smaller31, confirming the importance of current shunting to inactive neurons for coincidence detection in the hub-and-spoke circuit.
Figure 2.
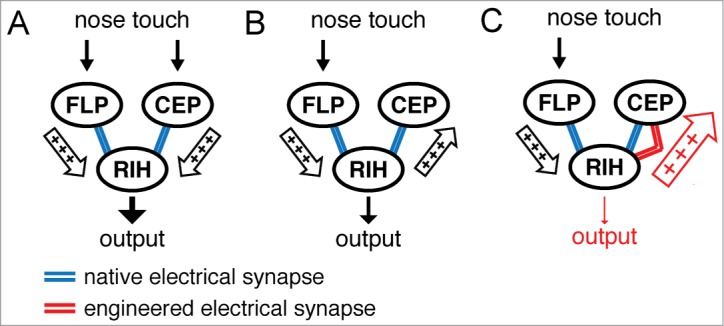
Enhancing electrical coupling in the nose touch circuit to increase shunting inhibition. (A) The nose touch circuit consists of several sensory neurons including FLP and CEP, which are each connected via electrical synapses to interneuron RIH. (B) When not all sensory neurons are activated the resulting circuit output as measured in RIH drops considerably, presumably due to current being shunted away from RIH into the inactive sensory neuron (e.g., CEP). (C) Artificially inserting a Cx36 electrical synapse between RIH and CEP further reduces the circuit output due to a larger shunting inhibition.31
Inserting novel electrical synapses between unconnected or chemically connected neurons
We also wished to examine whether ectopic electrical synapses could be introduced between uncoupled neurons or between neurons that are naturally connected by chemical synapses only. In this case, it would be possible to introduce not just a quantitative change to the weighting of the existing synaptic connectivity, but qualitatively modify the connectome by adding new connections.
We first considered the salt sensing neurons ASEL and ASER. The original C. elegans wiring diagram showed no chemical or electrical synapses to exist between these neurons4,5, and although more recent online data based on computer-aided reconstructions32 (http://wormwiring.org/) suggest some chemical connections may exist, these don't seem to be significant for salt sensing since the neurons respond to salt stimuli cell-autonomously33. ASEL and ASER show opposite responses to increases or decreases in salt concentration33 (Fig. 3A, left), which together shift the balance between the time spent moving forward and reorienting, ultimately producing net migration toward sources of moderately concentrated salt33. The processes of ASEL and ASER lie in close proximity to each other in the nerve ring. We therefore attempted to electrically couple these uncoupled neurons by inserting an electrical synapse between them. Following Cx36 expression in both neurons (Fig. 3A, middle) their calcium responses to salt presentation and removal changed dramatically22. For example, salt removal, which normally does not elicit a response in ASEL, produced an increase in ASEL calcium levels22 (Fig. 3A, right). This is consistent with positive charge flowing through an inserted electrical synapse from ASER to ASEL (Fig. 3A, middle). We were thus able to introduce a qualitative modification to the C. elegans connectome and add into it an otherwise non-existent electrical synaptic connection.
Figure 3.
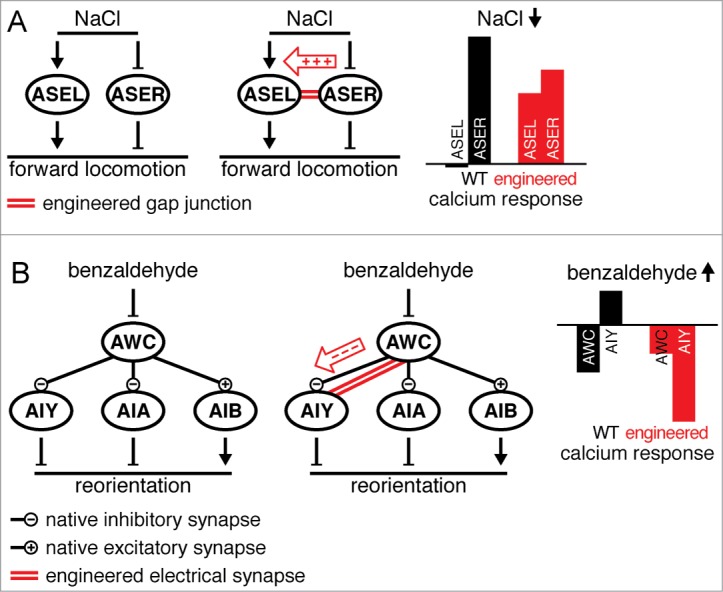
Inserting an electrical synapse between neurons that are not naturally connected by electrical synapses. (A) Salt sensing neuron ASEL and ASER act independently to transduce salt sensation (left). A Cx36 electrical synapse can be inserted between ASEL and ASER (middle). As a result, a decrease in salt concentration, which does not normally produce a response in ASEL, elicits a calcium increase in ASEL (right)22. (B) The olfactory circuit consists of sensory neuron AWC and its downstream chemical synaptic partners interneurons AIY, AIA and AIB (left). Inserting an otherwise non-existent electrical synapse between AWC and AIY (middle) flips the AIY response to increases in benzaldehyde concentration from positive to negative, switching its intrinsic anti-correlation with AWC into correlation (right).22
We also wished to apply this technique to modify the function of the olfactory circuit (Fig. 3B, left). The basic components of this circuit are the olfactory sensory neuron AWC and downstream interneurons AIY, AIA and AIB34,35. Increases in the concentration of attractants such as benzaldehyde reduce AWC activity, whereas decreases cause an increase in AWC activity. When AWC is depolarized it inhibits AIY and AIA and excites AIB through chemical synaptic transmission (Fig. 3B, left). The inhibition or excitation of these interneurons controls locomotion, ultimately guiding the worm toward the source of the attractive odor34,35. We inserted an electrical synapse between AWC and AIY (Fig. 3B, middle). The result, determined by calcium imaging, was a dramatic flip in the response properties of AIY from anti-correlation with AWC to correlation22. For example, following odor presentation, decreases in AWC activity, which normally entail no chemical synaptic transmission, produced an artificial decrease in AIY activity (Fig. 3B, right), presumably due to negative charge flowing from AWC into AIY through the new electrical synapse (Fig. 3B, middle). Although AIY is not the only interneuron in this circuit, the transmission of inverted information into it was sufficient to completely disrupt chemotaxis22. Interestingly, inserting an electrical synapse between AWC and AIA did more than abolish chemotaxis, it switched the response to benzaldehyde from attraction to repulsion (I.R. and W.R.S. unpublished data). Connecting between AWC and AIB enhanced the natural excitatory transmission between these 2 neurons (I.R. and W.R.S. unpublished data). Thus, engineered electrical connections can be integrated into existing neural circuits, reprogram their function and change the way they control behavior.
C. elegans as a prototype for connectome engineering
The long completed C. elegans connectome project and its important contributions might be considered as a pilot for current large-scale successive connectome projects. In a similar vein, the concepts behind the methodologies used for inserting new synapses into C. elegans neural circuits might be applicable for engineering other connectomes such as the fly (which also lacks endogenous connexins) or the mouse (where innexins rather than connexins could be ectopically expressed36). By pioneering the use of connectome editing in C. elegans, we hope to eventually lay the foundations for synthetic neuroscience in many other organisms.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. DeFelipe J. From the connectome to the synaptome: an epic love story. Science 2010; 330:1198-201; PMID:21109663; http://dx.doi.org/ 10.1126/science.1193378 [DOI] [PubMed] [Google Scholar]
- 2. Helmstaedter M. Cellular-resolution connectomics: challenges of dense neural circuit reconstruction. Nat Methods 2013; 10:501-7; PMID:23722209; http://dx.doi.org/ 10.1038/nmeth.2476 [DOI] [PubMed] [Google Scholar]
- 3. Van Essen DC. Cartography and connectomes. Neuron 2013; 80:775-90; PMID:24183027; http://dx.doi.org/ 10.1016/j.neuron.2013.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 1986; 314:1-340; PMID:22462104; http://dx.doi.org/ 10.1098/rstb.1986.0056 [DOI] [PubMed] [Google Scholar]
- 5. Varshney LR, Chen BL, Paniagua E, Hall DH, Chklovskii DB. Structural properties of the caenorhabditis elegans neuronal network. PLoS Comput Biol 2011; 7:e1001066; PMID:21304930; http://dx.doi.org/ 10.1371/journal.pcbi.1001066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bargmann CI, Marder E. From the connectome to brain function. Nat Methods 2013; 10:483-90; PMID:23866325; http://dx.doi.org/ 10.1038/nmeth.2451 [DOI] [PubMed] [Google Scholar]
- 7. Schafer WR. Deciphering the neural and molecular mechanisms of C. elegans behavior. Curr Biol 2005; 15:R723-9; PMID:16139205; http://dx.doi.org/ 10.1016/j.cub.2005.08.020 [DOI] [PubMed] [Google Scholar]
- 8. Michael G. What exactly is synthetic biology? Curr Biol 2011; 21:R611-4; http://dx.doi.org/ 10.1016/j.cub.2011.08.002 [DOI] [Google Scholar]
- 9. Mukherji S, van Oudenaarden A. Synthetic biology: understanding biological design from synthetic circuits. Nat Rev Genet 2009; 10:859-71; PMID:19898500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA. Rewiring cells: Synthetic biology as a tool to interrogate the organizational principles of living systems. Annu Rev Biophys 2010; 39:515-37; PMID:20192780; http://dx.doi.org/ 10.1146/annurev.biophys.050708.133652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Way JC, Collins JJ, Keasling JD, Silver PA. Integrating biological redesign: where synthetic biology came from and where It needs to go. Cell 2014; 157:151-61; PMID:24679533; http://dx.doi.org/ 10.1016/j.cell.2014.02.039 [DOI] [PubMed] [Google Scholar]
- 12. Miesenböck G, Kevrekidis IG. Optical imaging and control of genetically designated neurons in functioning circuits. Annu Rev Neurosci 2005; 28:533-63; PMID:16022604; http://dx.doi.org/ 10.1146/annurev.neuro.28.051804.101610 [DOI] [PubMed] [Google Scholar]
- 13. Deisseroth K. Optogenetics. Nat Meth 2011; 8:26-9; http://dx.doi.org/ 10.1038/nmeth.f.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tye KM. Neural circuit reprogramming: a new paradigm for treating neuropsychiatric disease? Neuron 2014; 83:1259-61; PMID:25233309; http://dx.doi.org/ 10.1016/j.neuron.2014.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pascoli V, Turiault M, Lüscher C. Reversal of cocaine-evoked synaptic potentiation resets drug-induced adaptive behaviour. Nature 2012; 481:71-5; http://dx.doi.org/ 10.1038/nature10709 [DOI] [PubMed] [Google Scholar]
- 16. Nabavi S, Fox R, Proulx CD, Lin JY, Tsien RY, Malinow R. Engineering a memory with LTD and LTP. Nature [Internet] 2014; 511:348-52 [cited 2014 Oct 27]; advance online publication. Available from: http://www.nature.com/nature/journal/vaop/ncurrent/full/nature13294.html; PMID:24896183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Daoudal G, Debanne D. Long-term plasticity of intrinsic excitability: learning rules and mechanisms. Learn Mem 2003; 10:456-65; PMID:14657257; http://dx.doi.org/ 10.1101/lm.64103 [DOI] [PubMed] [Google Scholar]
- 18. Zhang W, Linden DJ. The other side of the engram: experience-driven changes in neuronal intrinsic excitability. Nat Rev Neurosci 2003; 4:885-900; PMID:14595400; http://dx.doi.org/ 10.1038/nrn1248 [DOI] [PubMed] [Google Scholar]
- 19. Kato HK, Watabe AM, Manabe T. Non-hebbian synaptic plasticity induced by repetitive postsynaptic action potentials. J Neurosci 2009; 29:11153-60; PMID:19741122; http://dx.doi.org/ 10.1523/JNEUROSCI.5881-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sieber AR, Min R, Nevian T. Non-hebbian long-term potentiation of inhibitory synapses in the thalamus. J Neurosci 2013; 33:15675-85; PMID:24089475; http://dx.doi.org/ 10.1523/JNEUROSCI.0247-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abbott LF, Nelson SB. Synaptic plasticity: taming the beast. Nat Neurosci 2000; 3:1178-83; PMID:11127835; http://dx.doi.org/ 10.1038/81453 [DOI] [PubMed] [Google Scholar]
- 22. Rabinowitch I, Chatzigeorgiou M, Zhao B, Treinin M, Schafer WR. Rewiring neural circuits by the insertion of ectopic electrical synapses in transgenic C. elegans. Nat Commun [Internet] 2014. [cited 2014 Oct 27]; 5:4442. Available from: http://www.nature.com/ncomms/2014/140716/ncomms5442/full/ncomms5442.html#ref19; PMID:25026983; http://dx.doi.org/ 10.1038/ncomms5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takamori S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P, et al. Molecular anatomy of a trafficking organelle. Cell 2006; 127:831-46; PMID:17110340; http://dx.doi.org/ 10.1016/j.cell.2006.10.030 [DOI] [PubMed] [Google Scholar]
- 24. Bayés A, Grant SGN. Neuroproteomics: understanding the molecular organization and complexity of the brain. Nat Rev Neurosci 2009; 10:635-46; http://dx.doi.org/ 10.1038/nrn2701 [DOI] [PubMed] [Google Scholar]
- 25. McCracken CB, Roberts DCS. Neuronal gap junctions: expression, function, and implications for behavior [Internet] In: Ronald J. Bradley RAH, and Peter Jenner, editor. International Review of Neurobiology. Academic Press; 2006. [cited 2014 Oct 27] page 125-51. Available from: http://www.sciencedirect.com/science/article/pii/S0074774206730045. [DOI] [PubMed] [Google Scholar]
- 26. Hervé J-C, Phelan P, Bruzzone R, White TW. Connexins, innexins and pannexins: bridging the communication gap. Biochim Biophys Acta BBA—Biomembr 2005; 1719:3-5; http://dx.doi.org/ 10.1016/j.bbamem.2005.11.013 [DOI] [PubMed] [Google Scholar]
- 27. Epstein ML, Gilula NB. A study of communication specificity between cells in culture. J Cell Biol 1977; 75:769-87; PMID:562887; http://dx.doi.org/ 10.1083/jcb.75.3.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Condorelli DF, Parenti R, Spinella F, Salinaro AT, Belluardo N, Cardile V, Cicirata F. Cloning of a new gap junction gene (Cx36) highly expressed in mammalian brain neurons. Eur J Neurosci 1998; 10:1202-8; PMID:9753189; http://dx.doi.org/ 10.1046/j.1460-9568.1998.00163.x [DOI] [PubMed] [Google Scholar]
- 29. Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, Bargmann CI. A hub-and-spoke circuit drives pheromone attraction and social behavior in C. elegans. Nature 2009; 458:1171-5; PMID:19349961; http://dx.doi.org/ 10.1038/nature07886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chatzigeorgiou M, Schafer WR. Lateral facilitation between primary mechanosensory neurons controls nose touch perception in C. elegans. Neuron 2011; 70:299-309; PMID:21521615; http://dx.doi.org/ 10.1016/j.neuron.2011.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rabinowitch I, Chatzigeorgiou M, Schafer WR. A gap junction circuit enhances processing of coincident mechanosensory inputs. Curr Biol 2013; 23:963-7; PMID:23707432; http://dx.doi.org/ 10.1016/j.cub.2013.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu M, Jarrell TA, Wang Y, Cook SJ, Hall DH, Emmons SW. Computer assisted assembly of connectomes from electron micrographs: application to caenorhabditis elegans. PLoS One 2013; 8:e54050; PMID:23342070; http://dx.doi.org/ 10.1371/journal.pone.0054050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suzuki H, Thiele TR, Faumont S, Ezcurra M, Lockery SR, Schafer WR. Functional asymmetry in Caenorhabditis elegans taste neurons and its computational role in chemotaxis. Nature 2008; 454:114-7; PMID:18596810; http://dx.doi.org/ 10.1038/nature06927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chalasani SH, Chronis N, Tsunozaki M, Gray JM, Ramot D, Goodman MB, Bargmann CI. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature 2007; 450:63-70; PMID:17972877; http://dx.doi.org/ 10.1038/nature06292 [DOI] [PubMed] [Google Scholar]
- 35. Tsunozaki M, Chalasani SH, Bargmann CI. A behavioral switch: cGMP and PKC signaling in olfactory neurons reverses odor preference in C. elegans. Neuron 2008; 59:959-71; PMID:18817734; http://dx.doi.org/ 10.1016/j.neuron.2008.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Firme CP, 3rd, Natan RG, Yazdani N, Macagno ER, Baker MW. Ectopic expression of select innexins in individual central neurons couples them to pre-existing neuronal or glial networks that express the same innexin. J Neurosci 2012; 32:14265-70; PMID:23055495; http://dx.doi.org/ 10.1523/JNEUROSCI.2693-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]


