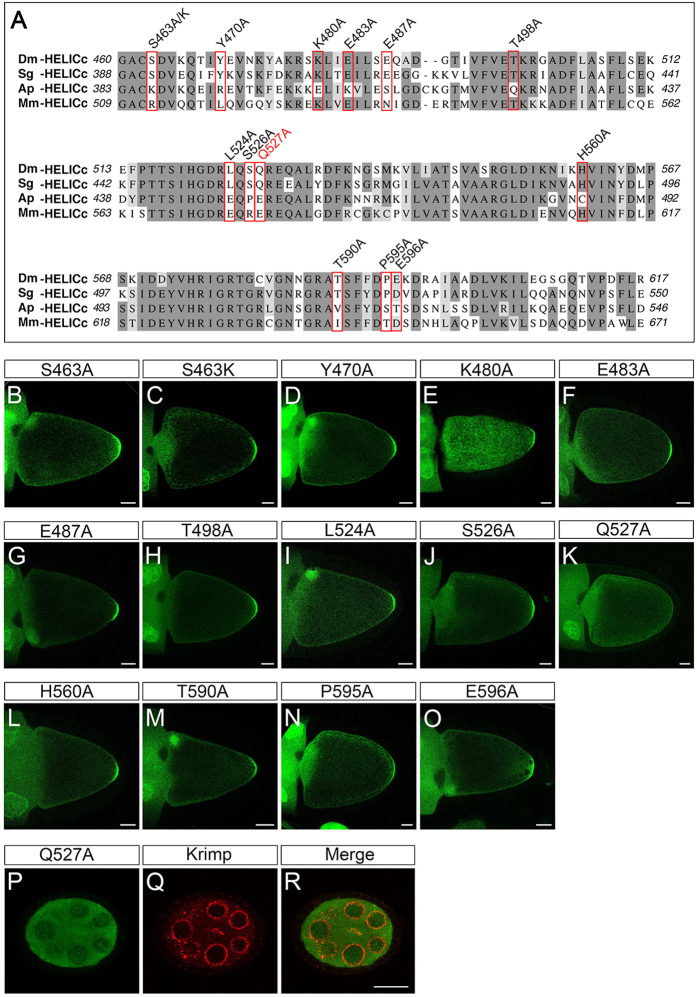
Figure 6. Identification of amino acid residues essential to the posterior localisation of Drosophila Vasa (DmVas) in the helicase superfamily C-terminal domain (HELICc).
(A) Multiple sequence alignment of HELICc domains belonging to Vas orthologs of Drosophila melanogaster (Dm), the grasshopper Schistocerca gregaria (Sg), the pea aphid Acyrthosiphon pisum (Ap), and the mouse Mus musculus (Mm). Dark grey: conserved residues, Light grey: residues with similar properties. Residues substituted by Ala or Lys for the localisation assays shown in panels (B–O) are highlighted with red boxes. (B–O) Localisation analysis of green fluorescent protein (GFP)-tagged DmVas460–661 proteins with replaced amino acid residues in the HELICc sequence. Stage-10 egg chambers were stained using the anti-GFP antibody (green). Anterior is to the left and posterior is to the right. Scale bars, 25 μm. (B) DmVas460–661/S463A: replacement of the Ser463 with Ala is designated as S463A, and this applies to the other replacements. (C) DmVas460–661/S463K. (D) DmVas460–661/Y470A. (E) DmVas460–661/K480A. (F) DmVas460–661/483A. (G) DmVas460–661/E487A. (H) DmVas460–661/T498A. (I) DmVas460–661/L524A. (J) DmVas460–661/S526A. (K) DmVas460–661/Q527A. (L) DmVas460–661/H560A. (M) DmVas460–661/T590A. (N) DmVas460–661/P595A. (O) DmVas460–661/E596A. All of the previously described DmVas460–661 variants could be localised to the germ plasm, except DmVas460–661/Q527A, shown in panel (K). (P–R) Stage-5 egg chambers expressing GFP-DmVas460–661/Q527A were double stained using the anti-GFP (green) and anti-Krimp antibodies (red). (P) GFP-DmVas460–661/Q527A was not colocalised with (Q) Krimp in the nurse cells. Anterior is to the left and posterior is to the right. Scale bars, 20 μm.