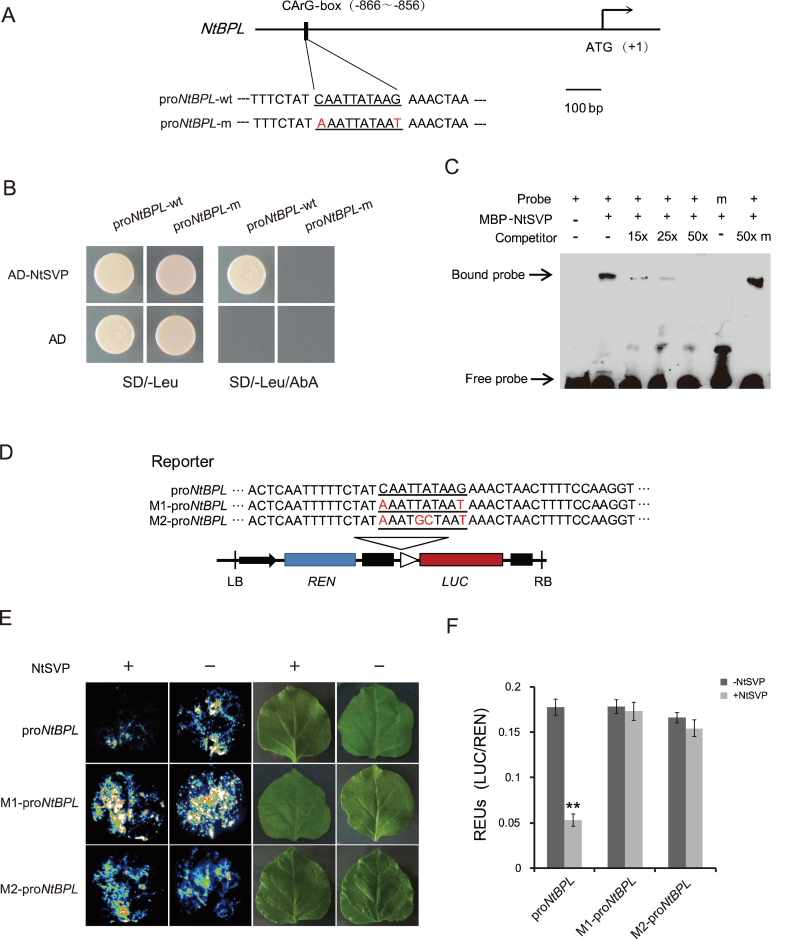
Fig. 4.
NtSVP as a direct transcription repressor of NtBPL. (A) The design of the wild-type (WT) probe containing the CArG-box (proNtBPL-wt) and the mutated probe (proNtBPL-m) in the NtBPL promoter. Mutated bases are highlighted in red. Numbers indicate sequence locations. (B) Yeast one-hybrid assays showing interactions between the NtSVP protein and the NtBPL promoter. pGADT7 (AD) was used as control. (C) EMSA showing MBP–NtSVP fusion protein binding to the probes of proNtBPL as described in (A). m represents proNtBPL-m as a control. + indicates the presence and – the absence of corresponding components as indicated. (D) The design of reporter constructs for the dual-luciferase assay. The WT and two mutant version of proNtBPL (M1, M2) are indicated. (E) LUC activities in N. benthamiana leaves infiltrated with the Agrobacterium strain harbouring the indicated reporter in the presence (+) or absence (–) of the co-transfected Agrobacterium strain harbouring the plasmid expressing the NtSVP protein. (F) Dual-luciferase assay of relative reporter activity of samples shown in (E). The relative LUC activities were normalized to REN activity and presented as relative expression units (REUs, n=5). **P<0.01.