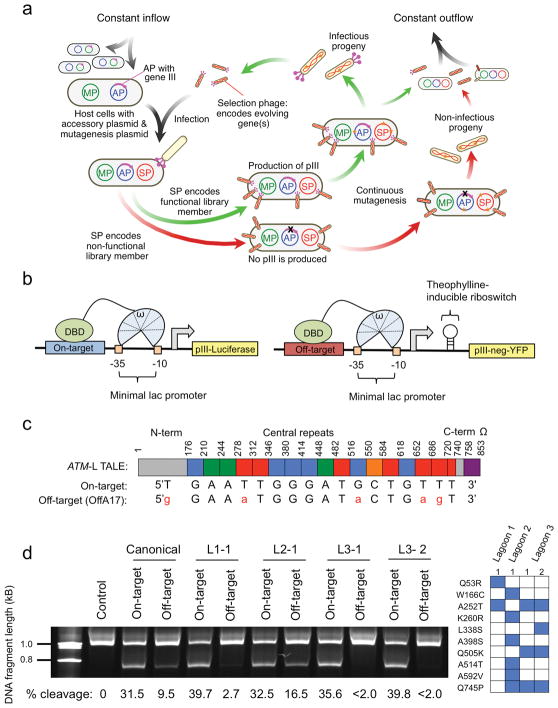
Figure 1. Development of DB-PACE and its application to TALEs.
(a) Overview of phage-assisted continuous evolution (PACE). (b) Left: Reporter system used to couple DNA binding to production of pIII-luciferase, and Right: Reporter system used to couple DNA binding to an off-target sequence to production of pIII-neg-YFP. (c) Schematic of the ATM-targeting TALE–ω fusion and the relationship between individual TALE repeats and the nucleotides they recognize for the on-target sequence (ATM: 5′-TGAATTGGGATGCTGTTT-3′), or the most highly cleaved human genomic off-target sequence (OffA17: 5′-GGAAATGGGATACTGAGT-3′). (d) Left: relative cleavage efficiencies of the canonical ATM TALEN pair or four ATM TALEN pairs containing the canonical ATM-right half site TALEN and an evolved ATM-left half site TALEN (L1-2, L2-1, L3-1, or L3-2) on a linear 6-kb DNA fragment containing either the ATM on-target sequence or the OffA17 off-target sequence. The top band is non-cleaved DNA, while the bottom band is a cleavage product. The gel has been cropped to simplify presentation. Cleavage percentages were determined using densitometry analysis (GelEval), and are included below each lane. Right: mutations in the evolved ATM-left half site TALEs used in the left panel.