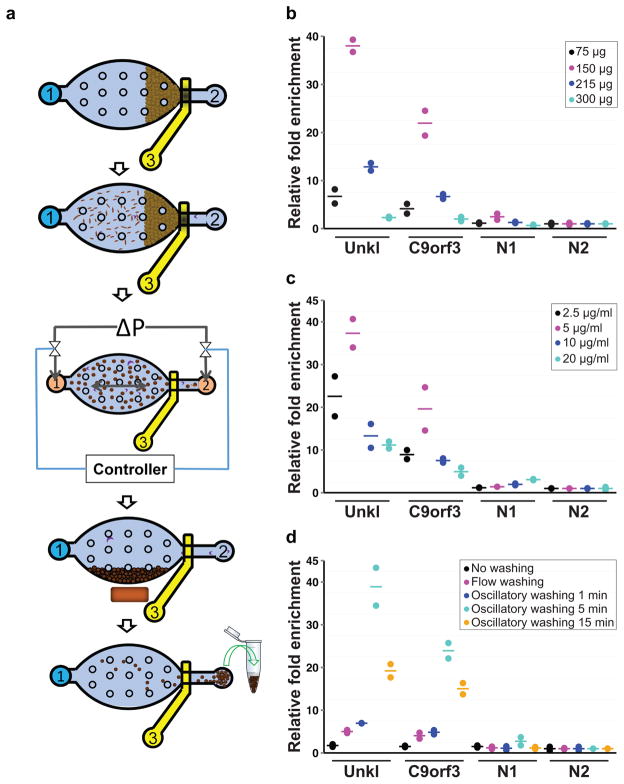
Figure 1. Overview of the MOWChIP-Seq protocol and its optimization.
(a) Schematic illustration for the five major steps of the protocol: step 1: Formation of a packed bed of IP beads; step 2: ChIP by flowing the chromatin fragments through the packed bed; step 3: Oscillatory washing; step 4: Removal of the unbound chromatin fragments and debris by flushing the chamber; step 5: Collection of the IP beads. The microfluidic chamber contains supporting pillars (shown as small circles) that prevent collapsing. (b–d) Optimization of the MOWChIP-Seq protocol. Major parameters of the protocol were optimized by checking for IP fold enrichment of known positive (UNKL and C9orf3) and negative loci (N1 and N2). IP was done against H3K4me3 in GM12878 cells. All experiments were conducted in duplicate and the horizontal lines represent the mean. Parameters optimized include: amount of beads in device chamber (b); concentration of antibody used for coating IP beads (c); washing duration in each of the two washing buffers (d). The relative fold enrichment was normalized against that of N2. 1000-cell samples were used in (b–d). 150 μg IP beads were used in (c, d). The antibody concentration for coating was 5 μg/ml for (b) and (d). The duration of oscillatory washing was 5 min for (b, c). Flow washing in (d) was implemented by flowing each washing buffer unidirectionally for 3 min under 1.5 μl/min.