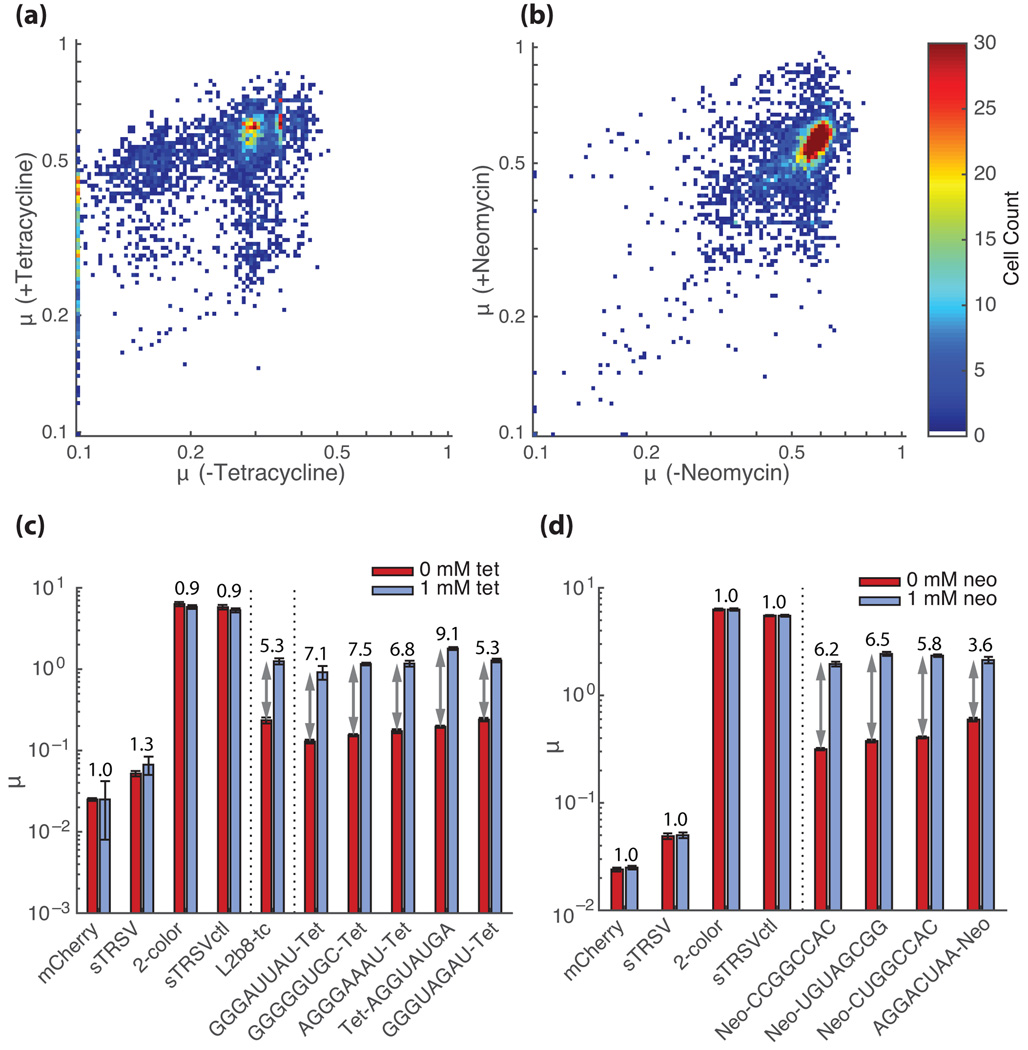
Fig. 4.
Extension of the FACS-Seq method to identifying tertiary interaction switches for other aptamer-target pairs. (a, b) GFP/mCherry ratios (µ) for tetracycline (a) and neomycin (b) aptamer library members in the presence and absence of target based on FACS-Seq assays. Each point on the plots represents a unique library sequence (tetracycline: N=3,873; neomycin: N=5,286; combined replicate data) that has the indicated (by color) µ under the specified conditions. Only sequences for which at least 50 cells were counted over the two combined replicates are used in each analysis. (c, d) Flow cytometry-based activity measurements for selected tetracycline-responsive (c) and neomycin-responsive (d) switches identified from the NGS analysis. Median GFP/mCherry ratios are reported for cells harboring the indicated switches and controls grown in the absence (red) and presence (blue; tetracycline: 1 mM, neomycin: 0.1 mM) of target. Error bars represent the standard error of the mean over at least three biological replicates. Activation ratios are reported above each construct. Vertical dotted lines separate ribozyme/controls, secondary structure switches, and tertiary structure switches. Additional compensation was performed for samples containing tetracycline (see Online Methods) due to fluorescence properties of this molecule.