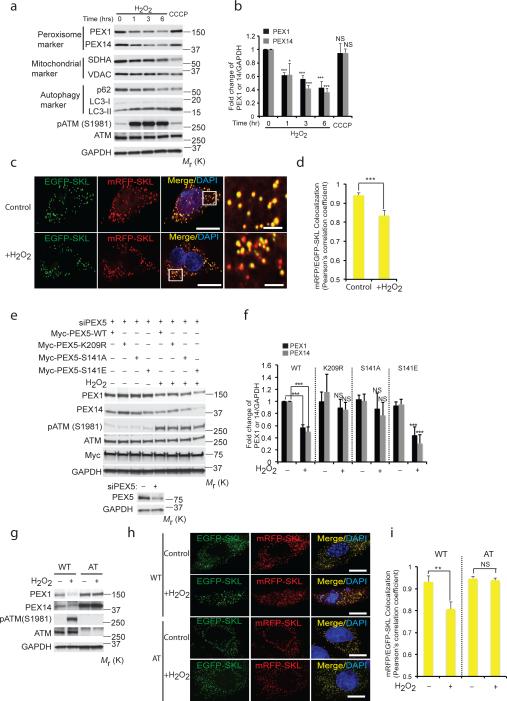
Figure 7. Induction of pexophagy by ROS.
(a) HepG2 cells treated with either H2O2 (0.4 mM) for indicated times or CCCP (50 μM, 6 h). Western analysis of peroxisome proteins PEX1 and PEX14 (SDHA and VDAC as mitochondrial markers, p62 and LC3-II as autophagy markers). (b) Quantification of PEX1 and PEX14 intensity normalized to GAPDH from Fig. 7a. (mean ± s.d., n = 3 independent experiments, student's t test). * P < 0.05 and *** P < 0.001, NS= not significant. (c) Representative images using HepG2 cells transfected with an mRFP-EGFP-SKL construct and treated with H2O2 for 6h. Scale bar, 10 μm. (d) Pearson's correlation coefficient for colocalization between mRFP-SKL and EGFP-SKL was calculated from Fig. 7c. n=4 independent experiment, 10 cells were analyzed in each experiments. All error bars represent s.d., (student's t test) *** P < 0.001. (e) HEK293 cells transfected with Myc-PEX5-WT or Myc-PEX5-K209R or Myc-PEX5-S141 for 24 h following prior siRNA knockdown of PEX5 for 48 h, treated with H2O2 (0.4 mM) for 6 h. Western analysis was performed using anti- PEX1, PEX14, phosphor-ATM (S1981), ATM, MYC and GAPDH antibodies. Corresponding immunoblots for HEK293 cells transfected with control or siRNA PEX5 showing levels of PEX5. (f) Quantification of PEX1 and PEX14 intensity normalized to GAPDH from Fig. 7e. (mean ± s.d., n = 4 independent experiments, student's t test). *** P < 0.001, NS= not significant. (g) WT (GM08399) and AT (GM05849) fibroblasts were treated with H2O2 (0.4 mM) for 6 h and immunoblotted with PEX1, PEX14, phospho-ATM, ATM and GAPDH antibodies. (h) Representative images using WT (GM08399) and AT (GM05849) fibroblasts transfected with a mRFP-EGFP-SKL construct and treated with H2O2 for 6h. Scale bar, 15 μm. (i) Pearson's correlation coefficient for colocalization between mRFP-SKL and EGFP-SKL was calculated from Fig. 7h. Quantification was performed from Fig S6a on n=3 independent experiments, 10 cells were analyzed in each experiment. All error bars represent s.d., (student's t test) ** P < 0.01, NS= not significant. Uncropped images of western blots are shown in Supplementary Fig. S9, Statistic source data for Fig. 7b,d,f and i can be found in Supplementary Table 1.