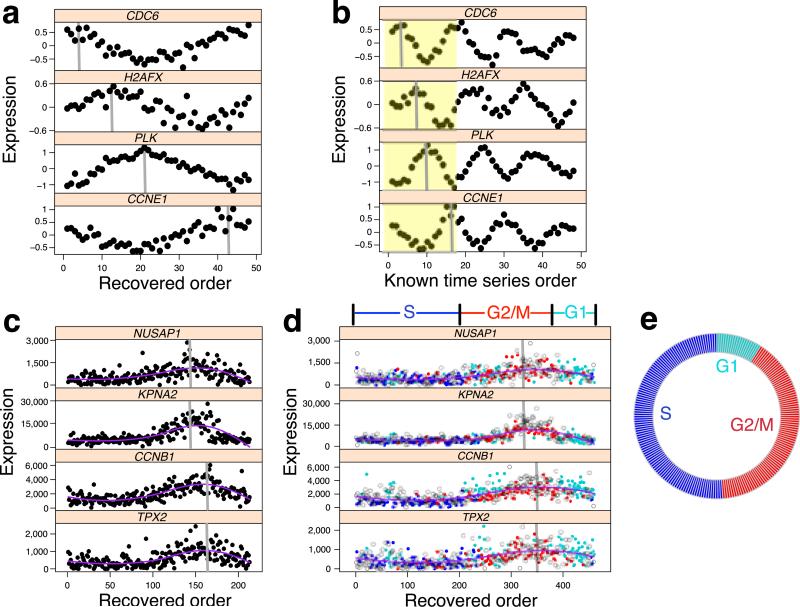
Figure 2.
Oscope uncovers oscillatory signals in case study datasets. (a) Four genes in the time series data from Whitfield et al. 20028 with profiles ordered by Oscope; the peak of the base cycle is marked in gray. (b) The same four genes following the known order over time with the peak of the first base cycle (shown in yellow) marked in gray. (c) Four genes from a 29 gene group identified by Oscope using scRNA-seq data from 213 unlabeled hESCs. Shown are Oscope recovered profiles. (d) The same four genes ordered using 460 cells (213 unlabeled and 247 H1-Fucci cells are shown as open circles and dots, respectively). The Fucci labels (ignored prior to applying Oscope) are shown in different colors for the 247 cells. Phase boundaries defined by the reconstructed order are shown above the plots. (e) The proportion of unlabeled cells that fall into each phase defined by the boundaries in d.