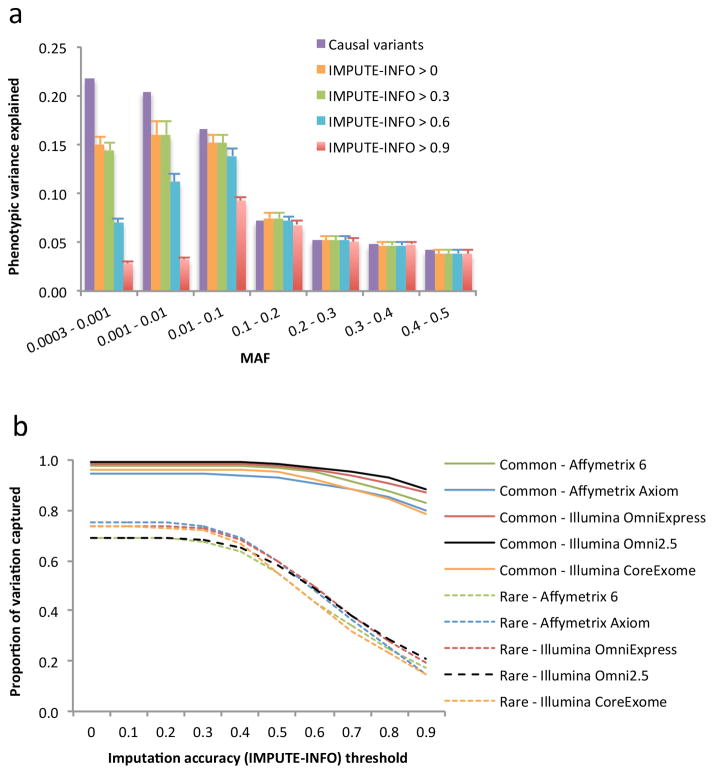
Figure 3.
Proportion of variation at sequence variants captured by 1KGP imputation in the UK10K-WGS data. The results are the averages from 200 simulations (Online Methods). Panel (a): estimates of proportion of phenotypic variance explained by 1KGP-imputed variants in different MAF groups from GREML-MS. The 1KGP imputation was based on variants on Illumina CoreExome array extracted from the UK10K-WGS data. The column in purple represents the variance explained by the causal variants. The other four columns represent the estimates using 1KGP-imputed variants filtered at 3 levels of imputation accuracy (IMPUTE-INFO) threshold. The error bar is the s.e.m.. Without filtering variants for IMPUTE-INFO (columns in yellow), the sum of the estimate is 96.2% for common variants and 73.4% for rare variants. Panel (b): estimates of proportion of variation at sequence variants captured by 1KGP imputation (the estimate of phenotypic variance explained by the 1KGP-imputed variants summed over MAF groups divided by that explained by the causal variants) based on different types of SNP genotyping arrays. Common: MAF > 0.01; Rare: 0.01 ≥ MAF > 0.0003.