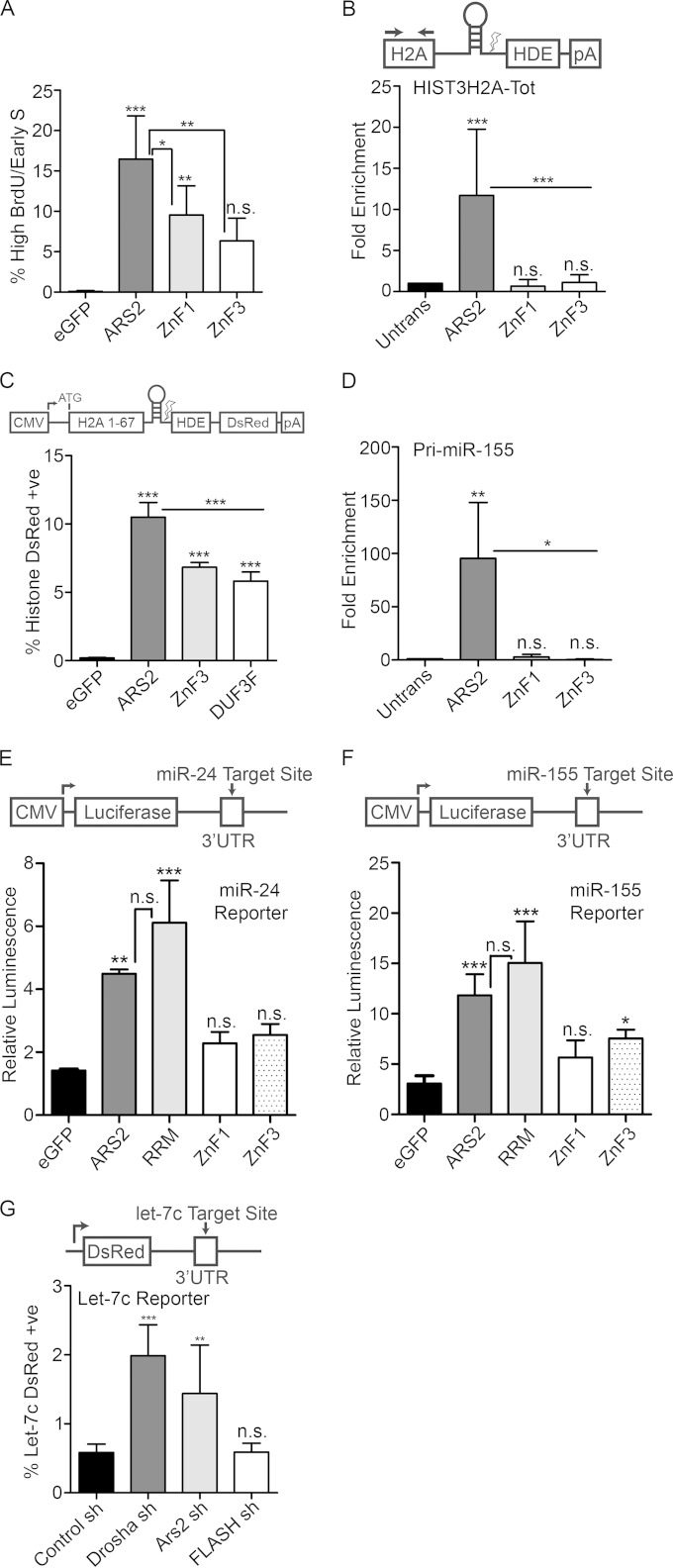
FIG 6.
The zinc finger domain mediates interaction with RNA. (A) Cells transfected with eGFP or eGFP-ARS2, -ZnF1 or -ZnF3 were pulsed with BrdU, fixed every 2.5 h, and gated for high GFP. Shown is the percentage of gated cells in the high-BrdU/early-S-phase-arrested population (average percent for each time point). (B) Cells transfected with 3×FLAG-ARS2 or the indicated mutants were immunoprecipitated, and the fold enrichment of RNA pulldown was quantified relative to that for untransfected samples using qPCR for HIST3H2A-Tot and primers upstream of the cleavage site. (C) Cells cotransfected with eGFP or eGFP-ARS2, -DUF3F, or -ZnF3 and histone DsRed were quantified as in Fig. 2E. (D) Samples were treated as for panel B and amplified using primers for pri-miR-155. (E and F) Cells were transfected with eGFP and eGFP-ARS2, -RRM, -ZnF1, or -ZnF3 along with the indicated firefly luciferase reporter and Renilla luciferase. Each firefly luciferase reporter contains an miRNA target site of perfect complementarity in its 3′ UTR. Firefly luminescence was normalized to Renilla luminescence. (G) Cells were cotransfected with shRNA containing a tGFP cassette and a let-7c DsRed reporter which contains a let-7c binding site in the 3′ UTR. The percent double positive (DsRed and GFP) is shown relative to total transfected cells. A one-way ANOVA was performed for panels A to G, with results as follows: F(3, 16) = 18.66 and P < 0.001 (A), F(3, 18) = 11.28 and P < 0.001 (B), F(3, 16) = 210.4 and P < 0.001 (C), F(3, 8) = 9.65 and P< 0.01 (D), F (4, 10) = 26.18 and P < 0.001 (E), F(4, 25) = 26.90 and P < 0.001 (F), and F(3, 20) = 22.84 and P < 0.001 (G), followed by Tukey's multiple-comparison post hoc test to determine whether there were significant differences between groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant. Error bars represent SD.