FIG 7.
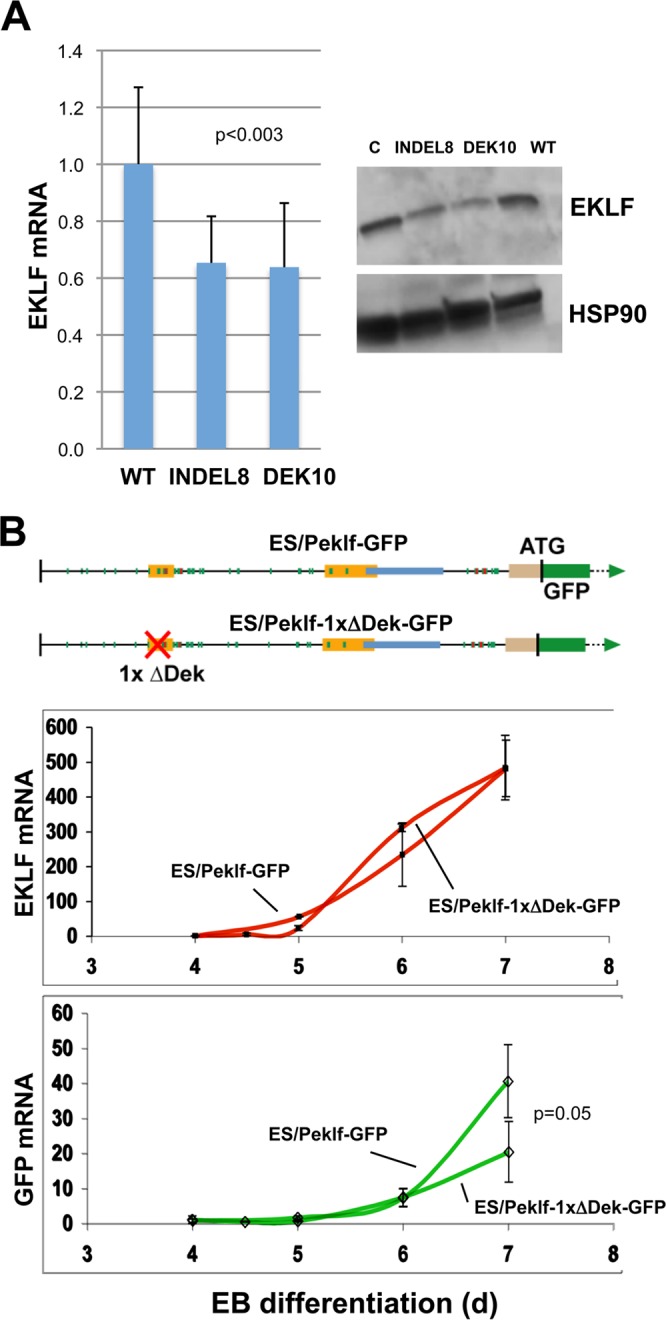
Importance of the oligonucleotide 2 sequence for optimal EKLF expression. (A) Total RNA was isolated or whole-cell extracts were prepared from wild-type (WT), indel8, or DEK10 MEL cells and analyzed for expression of EKLF RNA (quantitative RT-PCR; analysis of 11 analytical replicates each from two experiments) or protein (Western blotting) as indicated. “C” is an additional loading control to balance the EKLF protein signals across the gel. (B) (Top) Schematic of constructs containing the KLF1 promoter or one that has the oligonucleotide 2 sequence removed that were stably integrated into mouse ES cells adjacent to a GFP reporter, yielding two stable ES lines (“Peklf-GFP” and “Peklf-1xΔDek-GFP”). These are single-copy, unidirectional integrations into the same homing site, which thus avoids position effect variegation. Locations of the mapped upstream enhancer, proximal promoter, and intronic enhancer are as indicated. (Middle and bottom) ES cells from each were differentiated to EBs for the indicated number of days, and samples were quantitatively analyzed for expression of endogenous KLF1 (red) or exogenous reporter (green). The data show that the two cell lines differentiated and expressed endogenous KLF1 normally but that the Peklf-1xΔDek-GFP line generated a significantly lower relative level of reporter than did the Peklf-GFP line. Data are from three experiments analyzed in triplicate.
