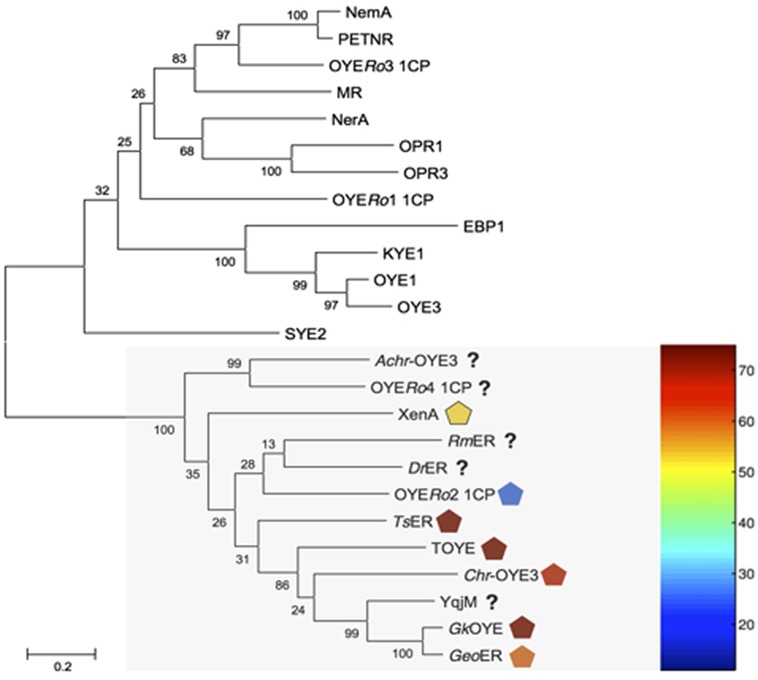
FIGURE 1.
Dendrogram of amino acid sequences of old yellow enzymes (OYEs) from Rhodococcus opacus 1CP and characterized OYE enzymes. The maximum likelihood distance tree was solved by usage of Mega 6.06-mac with Clustal W alignment method. Test of phylogeny was performed with 500 bootstrap replications, and resulting numbers are indicated. ‘Thermophilic-like’ OYEs are indicated by a light gray box. Melting temperatures of ‘thermophilic-like’ OYEs are indicated by symbols colored with respect to the thermogram. NCBI accession numbers are in parentheses. OYE1: Saccharomyces pastorianus (carlsbergensis, CAA37666), OYE3: S. cerevisiae (CAA97878), KYE1: Kluyveromyces lactis (AAA98815), EBP1: Candida albicans (AAA18013), OPR1: Solanum lycopersicum (CAB43506), OPR3: S. lycopersicum (CAC21424), NerA: Agrobacterium tumefaciens (CAA74280), MR: Pseudomonas putida (AAC43569), OYERo1 and OYERo3: R. opacus 1CP, NemA: Escherichia coli (BAA13186), PETNR: Enterobacter cloacae (AAB38638), SYE2: Shewanella oneidensis (AAN55487), Achr-OYE3: Achromobacter sp. JA81 (AFK73187), OYERo2 and OYERo4: R. opacus 1CP, RmER: Ralstonia (Cupriavidus) metallidurans CH34 (ABF11721), DrER: Deinococcus radiodurans R1 (AAF11740), TsER: Thermus scotoductus (CAP16804), XenA: P. putida (AAF02538), TOYE: Thermoanaerobacter pseudethanolicus (ABY93685), Chr-OYE3: Chryseobacterium sp. CA49 (AHV90721), YqjM: Bacillus subtilis (BAA12619), GkOYE: Geobacillus kaustophilus (BAD76617) and GeoER: Geobacillus sp. #30 (BAO37313).