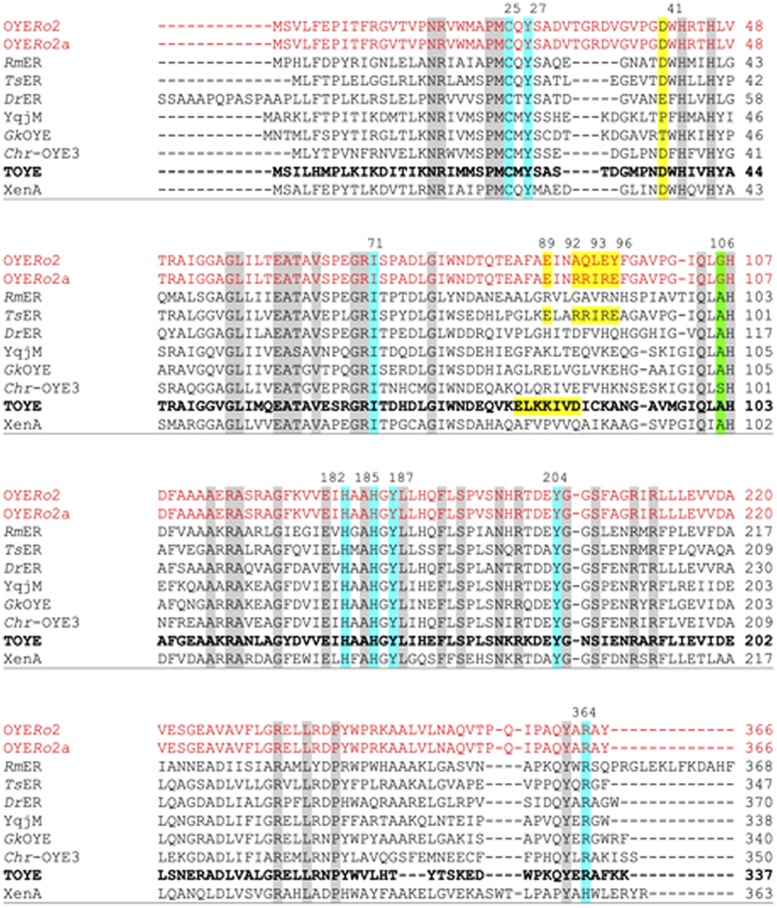
FIGURE 2.
Multiple sequence alignment of OYERo2 and OYERo2a with eight ‘thermophilic-like’ OYE homologs. Highly conserved residues are shaded in gray, conserved residues involved in the active site are shaded in blue and green. The salt bridges of TsER and TOYE are shaded in yellow. Amino acids that were replaced by site-directed mutagenesis of OYERo2 are also shaded in yellow in the wildtype and in the protein variant OYERo2a. Amino acid numbering of conserved residues is given according to the TOYE sequence (bold font). Other amino acid numberings are given according to each protein sequence. NCBI accession numbers and organism sources: RmER: R. (Cupriavidus) metallidurans CH34 (ABF11721), DrER: D. radiodurans R1 (AAF11740), TsER: T. scotoductus (CAP16804), XenA: P. putida (AAF02538), TOYE: T. pseudethanolicus (ABY93685), Chr-OYE3: Chryseobacterium sp. CA49 (AHV90721), YqjM: B. subtilis (BAA12619) and GkOYE: G. kaustophilus (BAD76617).