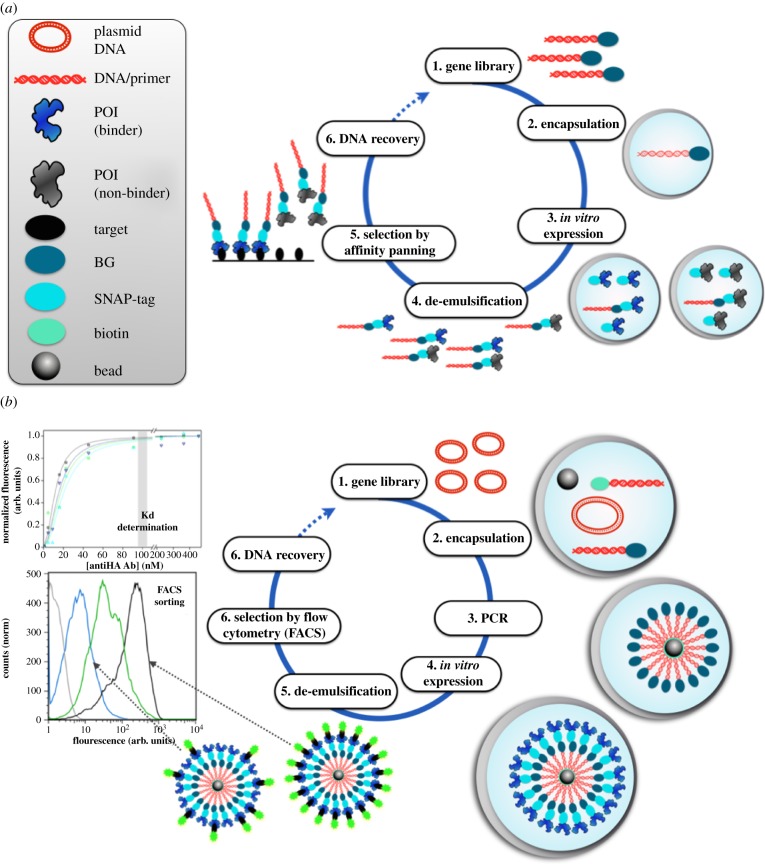
Figure 3.
Formats for artificial covalent genotype–phenotype linkages based on droplet compartmentalization. The key initial step of both display methods is that a DNA library (coding for SNAP-tag-fused variants of the POI) is compartmentalized in water-in-oil emulsion droplets, so that each compartment contains no more than one DNA template (Poisson distribution). (a) In SNAP display [29–33], the POI is in vitro expressed from a single gene in fusion to the SNAP-tag (1). The SNAP-tag of the expressed fusion protein then reacts with its substrate, BG, that has been covalently linked to the DNA template. As a result, the SNAP-tag connects genotype and the displayed protein (responsible for the phenotype). (2) SNAP-tagged proteins are de-emulsified and challenged for binding against an antigen by affinity panning. (3) After non-binders are washed away, binders are eluted together with their encoding genes that can feed the next round of selection. (b) In BeSD display [23], the DNA is amplified by ePCR (using appropriate labelled primers), captured on the beads via a biotin–streptavidin linkage and the POI is in vitro expressed. After the emulsion is broken, beads are incubated with the labelled target and the affinity for the target is measured via fluorescence-activated sorting (FACS). The binding affinity of each recovered variant can be measured by subsequent FACS analysis on the bead display construct. The bead connects genotype and the megavalently displayed protein (responsible for the phenotype).