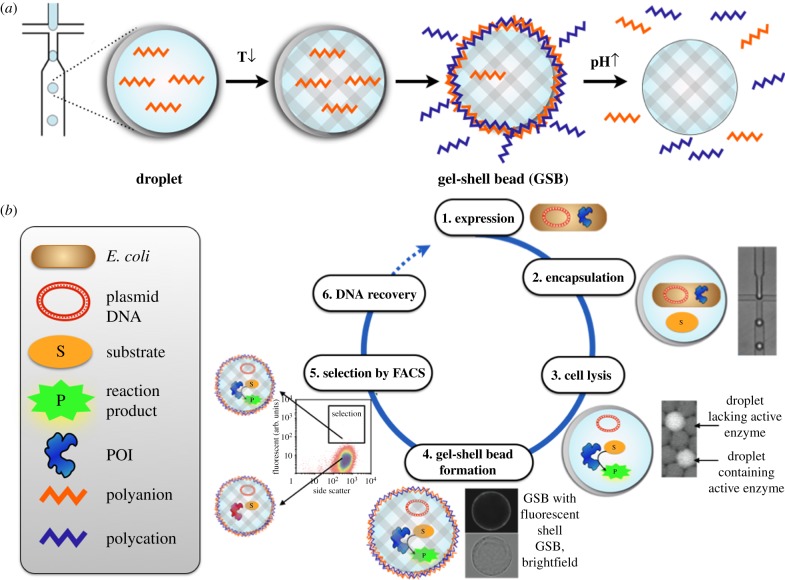
Figure 6.
GSBs for directed enzyme evolution [3]. (a) Assembly of GSBs: monodisperse water-in-oil emulsion droplets are produced with a microfluidic emulsion generator. The aqueous solution from which droplets are derived contains agarose and the polyanion alginate (red zig-zag line) that gelates upon cooling (from 37°C to 4°C), so that a solid bead is formed within the droplet template. The droplet boundary is removed by breaking the emulsion in the presence of the polycation poly(allylamine hydrochloride) (PAH; blue zig-zag line). Upon spontaneous encounter of the anionic alginate and the cationic PAH at the surface of the agarose template, a polyelectrolyte complex forms that maintains compartmentalization and substitutes the oil/water interface of the former emulsion droplet with a semipermeable surface layer, functionally resembling a semipermeable cell membrane. (b) Workflow of directed evolution in GSBs: (1) the protein-of-interest (POI), in this case an enzyme, is expressed in E. coli cells prior to droplet formation. (2) Single cells are then encapsulated into monodisperse water-in-oil emulsion droplets produced by a microfluidic emulsion generator (with Poisson distribution governing the occupancy). (3) Within the droplet, cells are lysed to allow encounter with reagents and substrate: now the enzymatic reaction can take place during an incubation period. (4) The gel-droplet emulsion is broken, or de-emulsified, in the presence of PAH to form the semipermeable polyelectrolyte shell layer, (5) GSBs harbouring active enzyme are distinguished from those with inactive enzyme variants by their fluorescence (and can be selected by flow cytometry, FACS). (6) The GSBs are broken up for genotype recovery.