Figure 1.
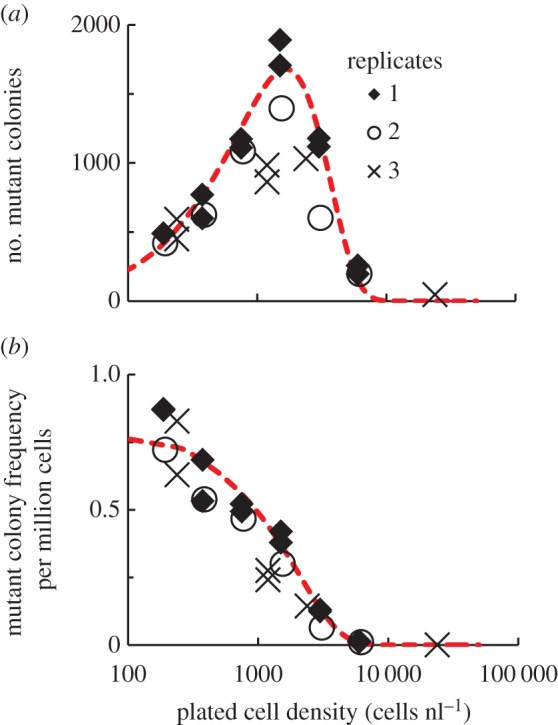
In a spatially mixed system, high cell densities negatively impact the survival of antibiotic-resistant mutants in the presence of antibiotic. Different cell densities from overnight PA14 cultures, containing WT cells and spontaneously generated antibiotic-resistant mutants, were plated on LB–tobramycin 4 μg ml−1 agar. (a) The number of growing antibiotic-resistant mutant colonies varies non-monotonically with cell density. n = 3; independent biological replicates are indicated by diamonds, circles and multiplication symbols. (b) The fraction of the plated culture that grows into antibiotic-resistant colonies decreases monotonically with increasing cell density. n = 3. In (a), the red, dashed line shows our analytical model, derived using a Poisson distribution to describe random fluctuations in density, that describes the number of surviving antibiotic-resistant mutants as a function of cell density. This model was fitted to the colony number data from replicate 1. R2 = 0.95. In (b), the red line shows the analytical model describing the frequency of mutant colonies as a function of cell density, using parameters determined from the previous fit. R2 = 0.78. Model parameters: Ve = 0.004, η = 1.164, Γ = 0.370 nlη−1 and μ = 1.10 × 10−6. (Online version in colour.)
