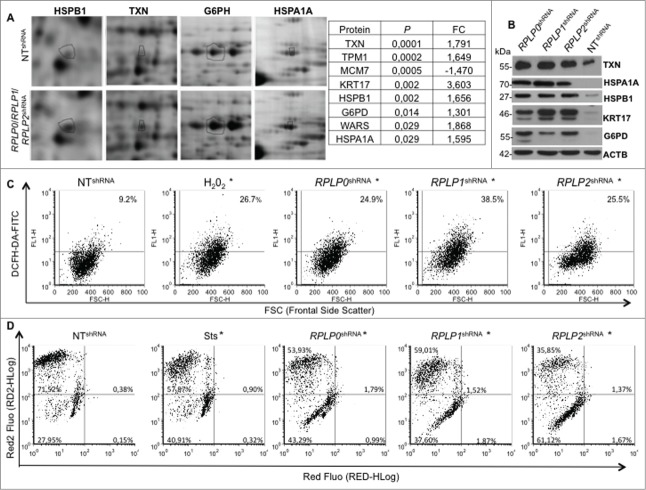
Figure 7.
Mitochondrial depolarization and redox state perturbed in MCF-7 cells stably expressing RPLP0 shRNA, RPLP1 shRNA, or RPLP2 shRNA vectors. (A) RPLP0 shRNA-, RPLP1 shRNA-, and RPLP2 shRNA-associated alterations in the levels of proteins that are upregulated by a factor of 1.5 or more in MCF-7 cells. Only portions of the 2D gel images are shown in each case (spots relevant to HSPB1, TXN, G6PH, and HSPA1A are highlighted with black circles). The right panel summarizes the proteins that were found to have the most significantly deregulated expression levels out of the 70 differentially expressed proteins (Table S1). (B) Western blotting analysis confirming the upregulation of the proteins identified in (A) as the most significantly deregulated proteins in MCF-7 cells expressing RPLP0 shRNA, RPLP1 shRNA, or RPLP2 shRNA with respect to controls expressing the NT shRNA vector. ACTB was used as a loading control. (C) Enhanced ROS generation after downregulation of RPLP0, RPLP1, or RPLP2 protein in MCF-7 cells. The percentages of DCFH-DA fluorescence represent the levels of intracellular O2−, as described in the Materials and Methods section. Parental MCF-7 cells treated with H2O2 were used as positive controls. Results were confirmed in 3 independent experiments (n = 3). Significant results were found for RPLP0, RPLP1, and RPLP2 deficient cells in comparison to control cells with *, P ≤ 0.05. (D) Flow cytometry-based evaluation of the change in the mitochondrial potential with the 7-AAD red fluorescent dye from the MitoPotential Red Kit. MCF-7 cells expressing RPLP0 shRNA, RPLP1 shRNA, RPLP2 shRNA, or NT shRNA vector and parental MCF-7 cells untreated or treated with 2 µM Sts for 3 h (positive control) were stained following the manufacturer's instructions, as described in the Materials and Methods. Quadrant gates were set up on the control cells and applied to the Sts-treated cells and cells expressing the different shRNAs. RPLP0 shRNA, RPLP1 shRNA, and RPLP2 shRNA samples are shown. The positive control treated with Sts underwent a change in mitochondrial potential that is shown in the graph as a downward shift in Red2 fluorescence and was quantified as the percentage of cells in the lower left quadrant. RPLP0 shRNA, RPLP1 shRNA, and RPLP2 shRNA samples do not show cell death, as shown by the few cells with an increase in red fluorescence. Results were confirmed in 3 independent experiments (n = 3). Significant results were found for RPLP0, RPLP1 and RPLP2 deficient cells in comparison to control cells with *, P ≤ 0.05.