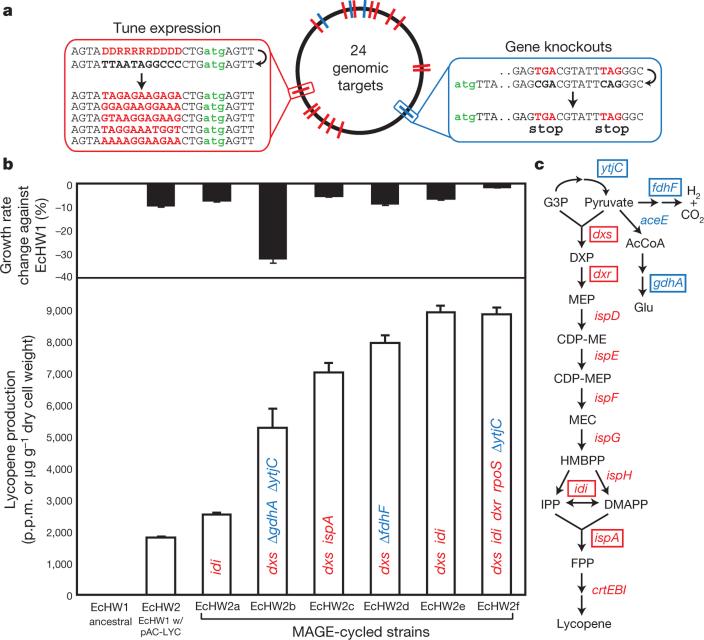
Figure 5. Optimization of the DXP biosynthesis pathway for lycopene production.
a, Genomic positions of 24 targeted genes with the RBS optimization strategy on the left (red) and gene knockout strategy on the right (blue). The gene knockout strategy involves the introduction of two nonsense mutations. All 90-mer oligos contain two phosphorothioated bases at the 3′ and 5′ termini. b, Black bars represent the growth rate of isolated variants (EcHW2a–f) relative to the ancestral EcHW1 strain. White bars represent lycopene production in p.p.m., which is normalized by dry cell weight in ancestral and mutant strains. Colour-coded labels in each white bar represent genetic modifications found by sequencing. All error bars indicate ± s.d.; n = 3. c, Modifications to the lycopene biosynthesis pathway of isolated variants EcHW2a–f with relevant genes highlighted by rectangular boxes. Blue labels represent knockout targets, red labels represent RBS tuning targets. AcCoA, acetyl-CoA; CDP-ME, 4-diphosphocytidyl-2-C-methyl-d-erythritol; CDP-MEP, 4-diphosphocytidyl-2C-methyl-d-erythritol-2-phosphate; DMAPP, dimethylallyl diphosphate; FPP, farnesyl diphosphate; G3P, glyceraldehyde 3-phosphate; HMBPP, (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate; IPP, isopentenyl diphosphate; MEP, 2-C-methyl-d-erythritol-4-phosphate; MEC, 2C-methyl-d-erythritol-2,4-cyclodiphosphate.