Fig 2. ChIP-seq enrichment represents genuine σ54 binding.
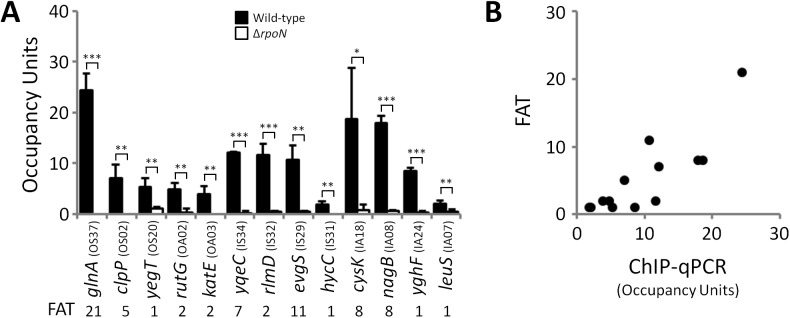
(A) Targeted validation of σ54 binding sites. ChIP-qPCR measurement of σ54 binding at putative sites identified by ChIP-seq in wild-type (MG1655; black bars) and ΔrpoN (RPB146; white bars) E. coli strains. The cognate promoter IDs and the fold above threshold (FAT; see Methods) scores (Tables 1 & 2) are indicated in parentheses and below the gene name, respectively. Gene names for “outside sense” (OS) and “outside antisense” (OA) binding sites correspond to the first gene downstream of the binding site (downstream relative to the orientation of the binding site). Gene names for “inside sense” (IS) and “inside antisense” (IA) binding sites correspond to the gene that contains the binding site. Occupancy units represent background-subtracted enrichment of target regions relative to a control region within the transcriptionally silent gene bglB. Error bars represent the standard deviation from three independent biological replicates. Significant differences between wild-type and ΔrpoN values are indicated (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001). (B) Correlation of ChIP-qPCR and ChIP-seq data. Values obtained from ChIP-qPCR (occupancy units) and ChIP-seq (FAT) using the σ54 antibody in E. coli were compared.
