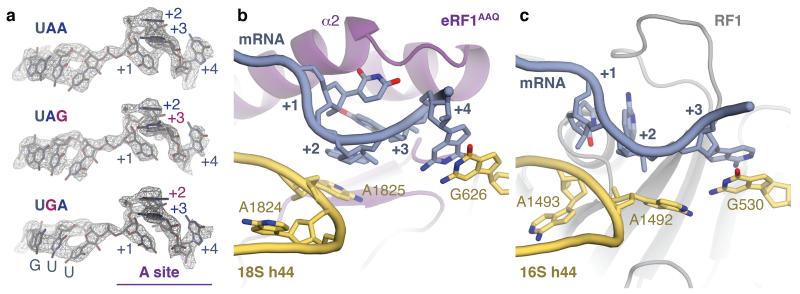
Figure 3. Stop codon configuration in the eukaryotic decoding centre.
a, EM map densities of the mRNA in the termination complexes containing the UAA, UAG and UGA stop codons reveal that they adopt the same compacted conformation. The ValGUU codon in the P site and the stop codon (+1 to +3) and following (+4) bases in the A site (purple) are indicated. b, The core termination signal recognised by eRF1AAQ (purple) is formed by four mRNA bases (+1 to +4, slate) that occupy the A site. Bases +2 and +3 stack on A1825, which is flipped out of helix 44 (h44), and base +4 on G626 of 18S rRNA (yellow). c, In bacteria, RF1 (grey) recognises a more extended stop codon configuration where the +3 base stacks on G530 (the equivalent of G626) of 16S rRNA.