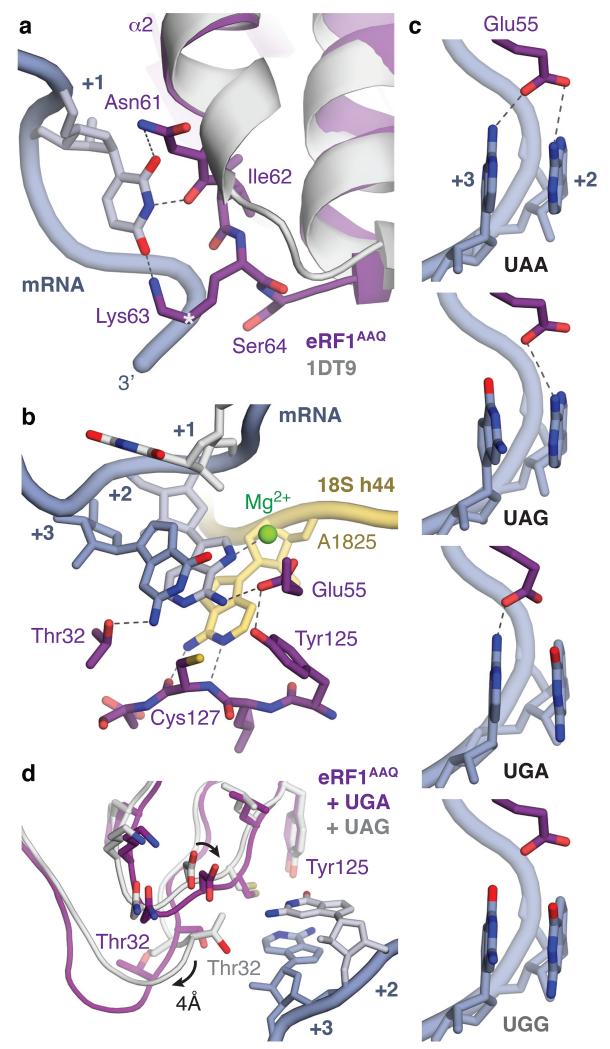
Figure 4. Molecular basis of stop codon recognition by eRF1.
a, The conformation of the NIKS motif (purple) at the end of helix α2 of ribosome-bound eRF1AAQ compared to the eRF1 crystal structure (PDB ID: 1DT9, light grey) allows it to form hydrogen bonds with the uridine in the +1 position of stop codons (slate; see also Extended Data Fig. 6). The hydroxyl groups of Ser64 and Cδ–hydroxylated (*) Lys63 help to position the NIKS motif by interacting with the mRNA phosphate backbone. b, Detailed interactions between the UAG stop codon (slate), eRF1AAQ (purple), and A1825 (yellow), depicting (i) stacking of the +2 and +3 bases with A1825, (ii) stabilisation of the flipped out position of A1825 by hydrogen bonds with the main chain of Cys127, (iii) a hydrogen bonding network involving Thr32, Glu55 and Tyr125 that specifies stop codon selectivity, and (iv) coordination of a Mg2+ ion (green) by the +2 adenosine. c, Model for stop codon discrimination by eRF1. Stacking of the +2 and +3 bases (slate) and possible hydrogen bonding interactions with Glu55 of eRF1 (purple) are shown for the three stop codons (UAA, UAG, UGA) and the sense codon UGG, which codes for tryptophan. d, The UGA stop codon induces a conformational change in the YxCxxxF and GTS motifs of eRF1AAQ (purple) compared to UAG- (white) and UAA-bound eRF1AAQ and eRF1 crystal structures.