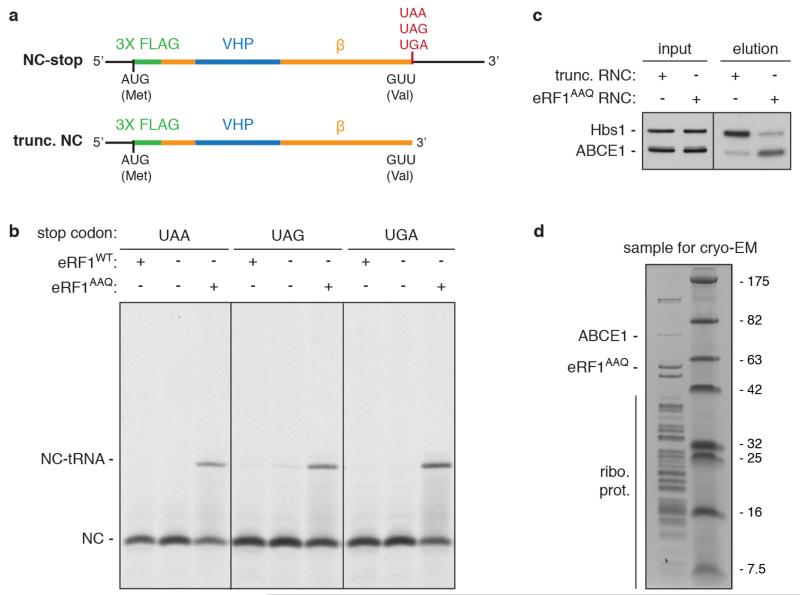
Extended Data Figure 1. eRF1AAQ stalls ribosomes at stop codons.
a, Line diagrams of mRNA encoding nascent chain (NC) substrates used in this study. The cytosolic portion of human Sec61β (residues 1-68, orange) was modified to contain an N-terminal 3× Flag tag (green) for affinity purification and the autonomously-folding villin headpiece (VHP, blue) domain. The three stop codons were individually inserted after Val68 of Sec61β to generate substrates for eRF1AAQ-mediated stalling, or the mRNA was truncated after the same residue to generate an independently-stalling substrate. b, In vitro translation reactions of NC-stop substrates containing the indicated stop codon (see panel a) in the presence of 35S-methionine without or with excess eRF1WT or eRF1AAQ. Reactions were for 25 min at 32°C and directly analyzed by SDS-PAGE and auto-radiography. The terminated (NC) and tRNA-associated (NC-tRNA) nascent chain products are indicated. Addition of eRF1AAQ selectively prevents peptide hydrolysis when the stop codon is reached. c, Anti-Flag affinity purifications of ribosome-nascent chains (RNCs) stalled either by mRNA truncation or at the UAA stop codon with eRF1AAQ (see panel a) were immunoblotted for the splitting factors Hbs1 and ABCE1. The different amounts of Hbs1 and ABCE1 co-purified despite identical nascent chain sequences in each RNC complex suggest that eRF1AAQ selectively traps ABCE1 on pre-termination complexes. d, SDS-PAGE and Coomassie staining of affinity-purified eRF1AAQ-stalled ribosome-nascent chains containing the UGA stop codon utilised for cryo-EM analysis. Bands corresponding to ribosomal proteins, ABCE1, and eRF1AAQ, which were verified by immunoblotting and mass spectrometry (data not shown), are indicated.