Fig. 5.
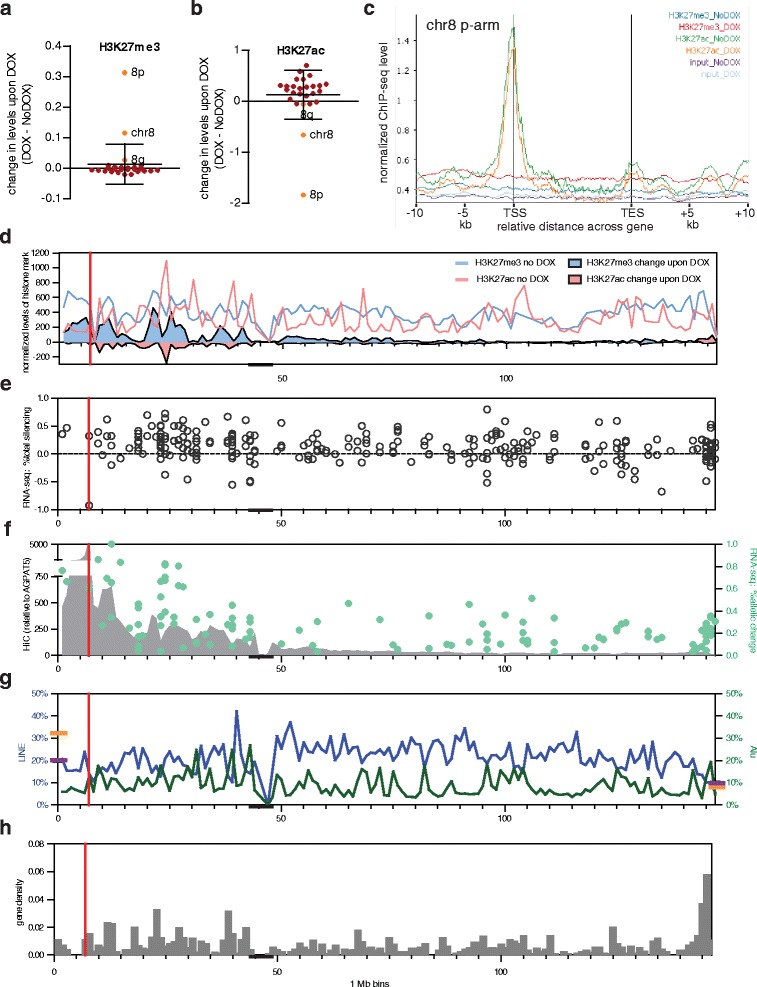
Correlation of genomic neighborhood and XIST-induced silencing on chromosome 8. a ChIP-seq changes observed for H3K27me3 on each chromosome, with chromosome 8 also subdivided into 8p (arm with integration) and 8q. H3K27me3 showed significant (P = 3.3e−243, paired t-test) changes for chromosome 8 with the genome showing no significant change. b ChIP-seq changes observed for H3K27ac on each chromosome, with chromosome 8 also subdivided into 8p (arm with integration) and 8q. The chromosome 8 decrease (P = 2.2e−40) as well as the genome-wide increase (P = 2.0e−205) were both highly significant. c Average H3K27ac and H3K27me3 normalized across genes on 8p. Normalized ChIP-seq level are shown across genes and for the 10 kb upstream and downstream of genes before XIST expression (NoDOX) and after XIST expression (DOX) as well as for input, color-coded as outlined. d Total H3K27me3 (blue line) and H3K27ac (pink line) in No DOX overlaid with change in H3K27me3 (blue), H3K27ac (pink) along chromosome 8 after 5 days of DOX induction of XIST. e Change in total reads for genes with average ≥5 FPKM shown as percent silencing. f Density of nuclear contacts with the 8p integration site (AGPAT5) as identified in IMR90 fibroblast cells by HiC [45] in 1 Mb bins (grey shading). Allelic silencing of genes is shown superimposed as green dots. g Density of LINE (blue line) and ALU (green line) repetitive elements per 1 Mb bin. Shown on the axes are the average genome (purple) and X-chromosomal (orange) densities of the elements. h Gene density in 1 Mb bins along the chromosome
