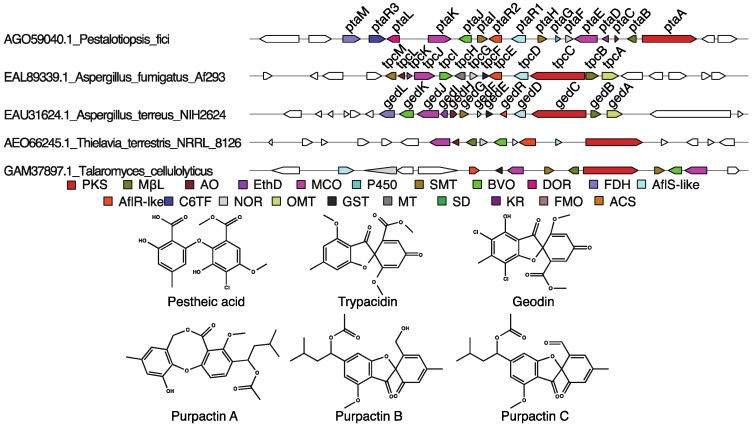
Figure 5.
Examples of tpc-like clusters of interest. At top, the gene cluster diagrams are shown for the characterized pta, tpc, and ged clusters as well as two uncharacterized gene clusters from the group V1 chosen from the larger group V phylogenetic tree, Figure S2. Genes are represented as arrows with a color corresponding to the proteins they encode which are detailed in the color key below the cluster diagrams. Genes with no color were not identified as homologous to any group V1 cluster gene. The structures of the characterized products from this clade, pestheic acid, trypacidin, and geodin, are shown below the color key. Below these are three metabolites reported to be produced by T. cellulolyticus (now considered synonymous with T. pinophilus), purpactins A–C [87]. PKS = Polyketide synthase, MβL = Metallo-β-lactamase-type thioesterase, AO = Anthrone oxidase, EthD = EthD domain-containing protein, a putative decarboxylase [51,53], MCO = multicopper oxidase, P450 = cytochrome P450, SMT = S-adenosylmethionine-dependent methyltransferase, BVO = Baeyer-Villiger oxidase, DOR = Pyridine nucleotide-disulfide oxidoreductase, FDH = Flavin-dependent halogenase, AflS = Transcriptional co-regulator of the aflatoxin biosynthetic gene cluster [76], AflR = Transcriptional regulator of the aflatoxin biosynthetic gene cluster, C6TF = GAL4-like Zn(II)2Cys6-domain and fungal-specific transcription factor domain-containing protein, NOR = NADH-dependent oxidoreductase, OMT = O-methyltransferase, GST = Glutathione S-transferase, MT = Methyltransferase, SD = Scytalone dehydratase, KR = Ver-1-like ketoreductase [77,78], FMO = Flavin-dependent monooxygenase, ACS = Acyl-CoA synthase.