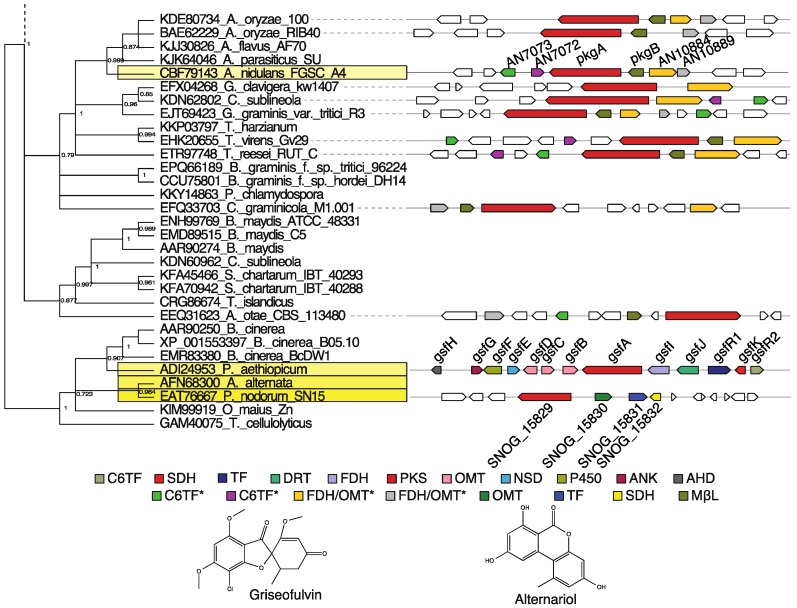
Figure 7.
A clade of gsf-, pkg-, PskI-, and SNOG_15820-like clusters. An excerpt of the group V phylogenetic tree made with FastTree [60], Figure S2, containing the PKSs from the griseofulvin-producing cluster, GsfA (ADI24953) [48], three alternariol-producing clusters [54,61,62], and a group of related uncharacterized PKSs, is shown at top left. The bootstrap values are presented next to their corresponding nodes. The yellow boxes indicate PKSs from the same species in which the characterized clusters were originally described. Next to the tree are the gene clusters corresponding to the PKSs that were identifiable through MultiGeneBLAST analysis. Genes are represented as arrows with a color corresponding to the proteins they encode which are detailed in the color key below the tree and cluster diagrams. Asterisks signify potential gene truncation due to misannotation. Genes with no color were not identified as homologous to any group V3 cluster gene. The products of the characterized examples from this clade, griseofulvin and alternariol, are shown at bottom. C6TF = GAL4-like Zn(II)2Cys6-domain and fungal-specific transcription factor domain-containing protein, GsfR2-like, SDH = Short chain dehydrogenase, TF = GAL4-like Zn(II)2Cys6-domain and fungal-specific transcription factor domain-containing protein, GsfR1-like, DRT = Drug resistance transporter, EmrB subfamily, FDH = Flavin-dependent halogenase, PKS = Polyketide synthase, OMT = O-methyltransferase, NSD = Nucleoside-diphosphate-sugar dehydratase, P450 = Cytochrome P450, ANK = Ankyrin repeat-containing protein, AHD = YcaC-related amidohydrolase, FDH/OMT = Flavin-dependent halogenase and O-methyltransferase bifunctional protein, MβL = Metallo-β-lactamase-type thioesterase. * Asterisks signify potential gene truncation due to misannotation.