Figure 1.
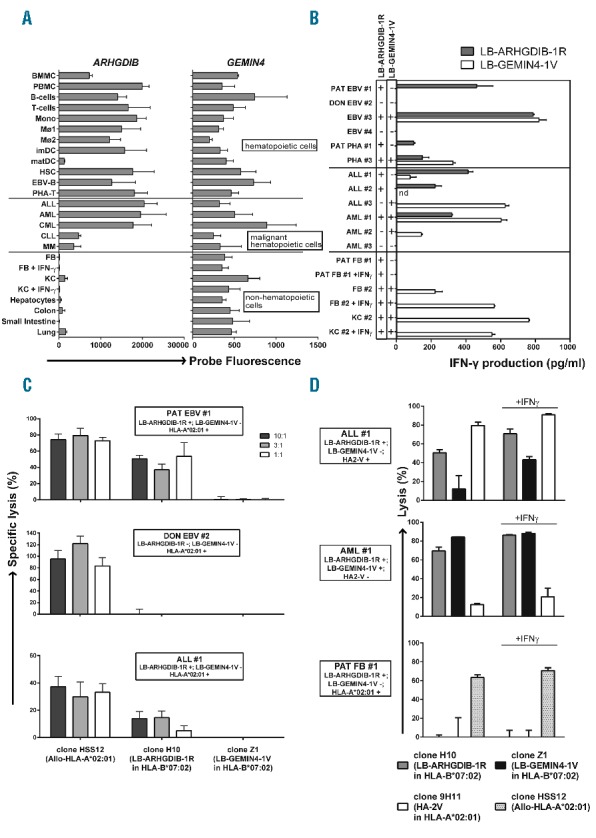
LB-ARHGDIB-1R as a target with therapeutic relevance for leukemia. (A) Expression profiles for ARHGDIB and GEMIN4 as determined by microarray gene expression analysis. GEMIN4 was selected as a representative gene for MiHA with ubiquitous expression on (non)-La by (non-)hematopoietic cells.5 The probe fluorescence, as measured on Illumina Human HT-12 v3/4 BeadChips, is indicated.8 Hematopoietic cells included bone marrow and peripheral blood mononuclear cells (BMMC and PBMC), B cells, T cells, monocytes (Mono), macrophages type I and II (MØ1 and MØ2), (im)mature DC (imDC and matDC), hematopoietic stem cells (HSC), Epstein-Barr virus-infected B (EBV-B) cells and phytohemagglutinin-stimulated T (PHA-T) cells. Malignant hematopoietic cells included acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic myeloid leukemia (CML), chronic lymphocytic leukemia (CLL) and multiple myeloma (MM). Non-hematopoietic cells included fibroblasts (FB) and keratinocytes (KC) cultured from skin biopsies in the absence or presence of IFN-γ and hepatocytes, colon and small intestine epithelial cells and lung epithelial cells. (B–D) Recognition and lysis of (non-)hematopoietic cells as mediated by T cells for LB-ARHGDIB-1R (clone H10).5 All samples were positive for HLA-B*07:02. Cell types included EBV-B and PHA-T cells from patient (PAT EBV #1 and PHA #1) or donor (DON EBV #2) origin or from third party individuals (EBV #3, EBV #4 and PHA #3) as well as primary ALL (ALL #1–3) and AML (AML #1–3) samples. One representative example of two independent experiments is shown. (B) Recognition of LB-ARHGDIB-1R in HLA-B*07:02 as expressed on (malignant) hematopoietic cells, but not on (IFN-γ pre-treated) FB and KC. Malignant cells were isolated by flow cytometry cell sorting based on CD19 (ALL) or CD33 (AML) expression. T cells for LB-GEMIN4-1V (clone Z1) were included as a positive control. Genotyping results (+ or −) for the single nucleotide polymorphisms encoding LB-ARHGDIB-1R (filled bars) and LB-GEMIN4-1V (open bars) are shown. Mean release of IFN-γ of duplicate wells as measured by ELISA after overnight co-incubation of T cells and stimulator cells at a ratio of 1:6 is depicted. (C) Cytolysis of primary leukemic blasts as mediated by T cells for LB-ARHGDIB-1R in a 51Cr-release assay. Target cells (1×103) labeled with Na251CrO4 (Perkin Elmer, Waltham, MA, USA) were co-incubated with T cells for 10 hours (h) at effector:target ratios of 10:1, 3:1 and 1:1. Mean specific lysis of triplicate wells is shown for patient and donor EBV-B (upper and middle graphs, respectively) and ALL #1 expressing LB-ARHGDIB-1R and HLA-B*07:02 (lower graph). Patient and donor EBV-B and ALL #1 were positive for HLA-A*02:01 and negative for LB-GEMIN4-1V. Allo-HLA-A*02:01 reactive T cells (clone HSS12) and T cells for LB-GEMIN4-1V were included as controls. (D) Cytolysis of primary leukemic blasts and patient’s FB as mediated by T cells for LB-ARHGDIB-1R in a 48 h flow cytometry-based cytotoxicity assay. T cells (2.5×104) were labeled with PKH67 (Sigma-Aldrich, St. Louis, MO, USA) and co-incubated for 48 h with AML or ALL samples or FB cultured from a skin biopsy of the patient in the absence or presence of IFN-γ (1×104). After co-incubation, cultures were stained with CD33-APC (AML), CD19-PE (ALL) or CD90-PE (FB). Sytox blue was added to gate on viable cells and flow count fluorospheres (Beckman Coulter, Brea, CA, USA) were added to calculate specific lysis. T cells recognizing HA-2 in HLA-A*02:01 (clone 9H11), T cells for LB-GEMIN4-1V and Allo-HLA-A*02:01 reactive T cells were included as controls. Mean lysis of triplicate wells is depicted.
