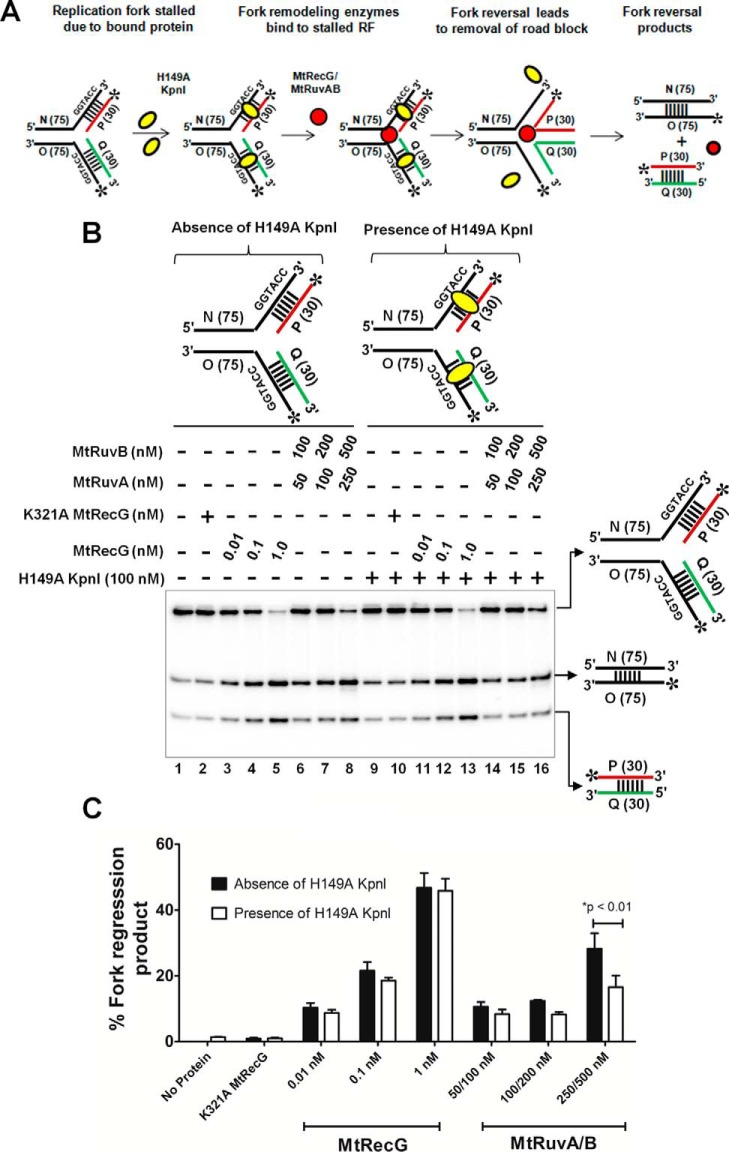
FIGURE 11.
M. tuberculosis RecG but not RuvAB drives efficient reversal of forks bound by protein. A, schematic representation of a fork bound by protein and the expected products after fork reversal reaction catalyzed by MtRecG and MtRuvAB. Asterisk indicates 32P label; oval represents KpnI H149A mutant; and dashed lines represent binding site for H149A KpnI enzyme. B, fork reversal activities of MtRecG and MtRuvAB with homologous replication fork structures either in the absence (lanes 1–8) or presence (lanes 9–16) of 100 nm H149A KpnI. Reactions were initiated by addition of indicated concentrations MtRecG (lanes 3–5 and 11–13) and MtRuvA + MtRuvB (lanes 6–8 and 14–16). Lanes 2 and 10 represent reactions with K321A MtRecG in the absence or presence of H149A KpnI. Reaction products were separated on 8% native polyacrylamide gel and analyzed by autoradiography. C, quantitative analysis of MtRecG- and MtRuvAB-driven fork reversal of homologous fork structures either bound by H149A KpnI or naked substrates. The error bars represent standard error of the mean (S.E.) from three independent experiments.