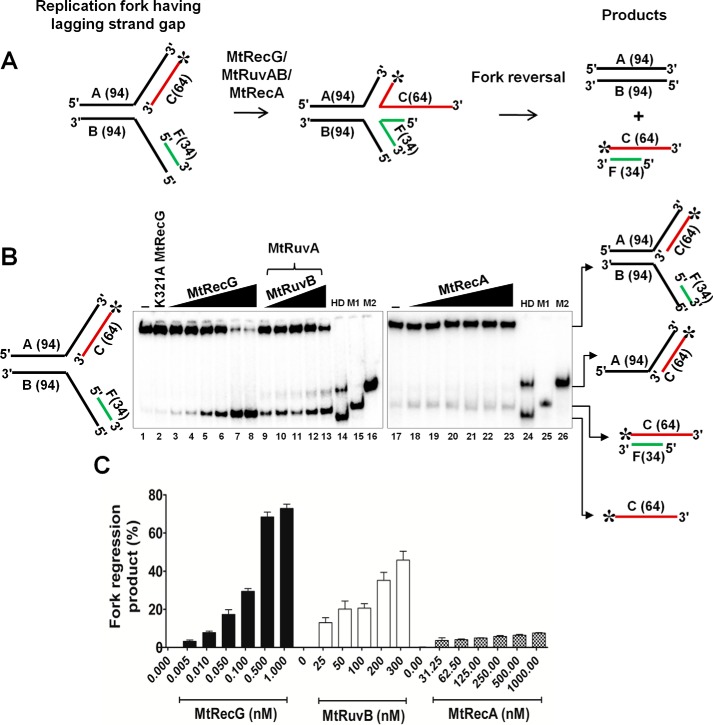
FIGURE 6.
Comparative RFR activities of M. tuberculosis RecG, RuvAB, and RecA on homologous replication fork containing lagging strand gap. A, schematic representation and outcome of RFR of homologous replication fork containing lagging strand gap. The asterisk indicates 32P label. B, analysis of RFR catalyzed by MtRecG, MtRuvAB, and MtRecA. Reaction mixture contained 10 nm 32P-labeled substrate in the presence of 1 mm ATP and 5 mm MgCl2. Reactions were initiated by addition of 0.005, 0.01, 0.05, 0.1, 0.5, and 1.0 nm MtRecG (lanes 3–8, respectively), 50 nm MtRuvA, and 25, 50, 100, 200, and 300 nm MtRuvB (lanes 9–13, respectively) and 31.25, 62.5, 125, 250, 500, and 1000 nm MtRecA (lanes 18–23, respectively). Lanes 14 and 24 represent heat-denatured substrate. Lanes 15 and 25 (M1) show a marker of leading and lagging strand annealed products, respectively. Lanes 16 and 26 (M2) represent a marker for products that are generated by helicase unwinding of the parental strands. Lanes 1 and 17 denote reactions in the absence of any protein. Lane 2 represents reaction with 10 nm K321A MtRecG. C, quantitative data for the MtRecG-, MtRuvAB-, and MtRecA-catalyzed RFR of substrates that contain lagging strand gap. The error bars represent standard error of the mean (S.E.) from three independent experiments.